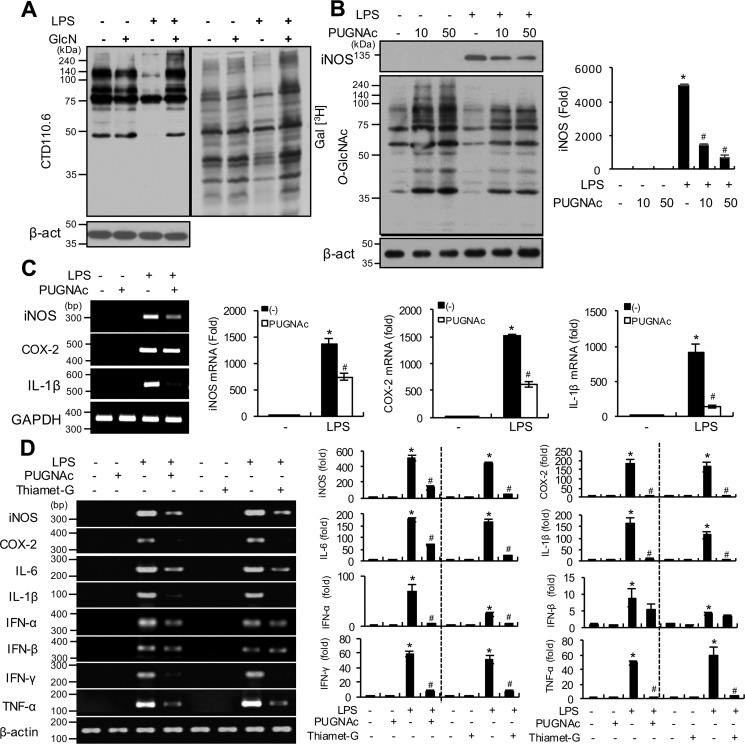
Figure 9.
O-GlcNAcylation changes in response to LPS and/or GlcN in RAW264.7 cells and effects of OGA inhibitors on LPS-induced inflammatory gene induction. A, RAW 264.7 cells were pre-treated with GlcN (5 mm) for 2 h, followed by stimulation with LPS (100 ng/ml) for 24 h. Whole cell lysates were prepared and subjected to Western blotting for O-GlcNAcylation using the CTD110.6 antibody (left panel) or galactosyltransferase-labeled using [3H]UDP-galactose as a substrate and exposed to X-ray film for autoradiography (right panel). B, RAW 264.7 cells were pre-treated with PUGNAc (10 or 50 μm) for 2 h and subsequently stimulated with LPS (100 ng/ml) for 24 h. O-GlcNAcylation levels of total cell lysates were measured via Western blotting using the CTD110.6 antibody. The graph represents relative densitometric intensities that were quantitatively measured and normalized to β-ACTIN levels. C, RAW 264.7 cells were stimulated with LPS (100 ng/ml) with or without PUGNAc (0.5 mm) for 24 h. Expression of iNos, Cox-2, and Il-1β mRNA were determined by RT-PCR (left panel) and quantitatively assessed using real-time PCR (right graphs). Gapdh was determined as the loading control. Results are representative of three independent experiments. All values are mean ± S.E. D, adult zebrafish were treated with PUGNAc (50 μg/g) or Thiamet-G (20 μg/g) for 2 h and injected with LPS (100 μg/g). Visceral tissues were isolated at 24 h and mRNA expression of inos, cox-2, il-6, il-1β, ifn-α, ifn-β, ifn-γ, and tnf-α determined using PCR (left panel) and quantitative real-time PCR (right graphs). Results are representative of three independent experiments. All values are mean ± S.E. Asterisk denotes significantly increased from the untreated control (p < 0.05); hash mark (#) indicates significantly decreased from LPS-stimulated conditions (p < 0.05).