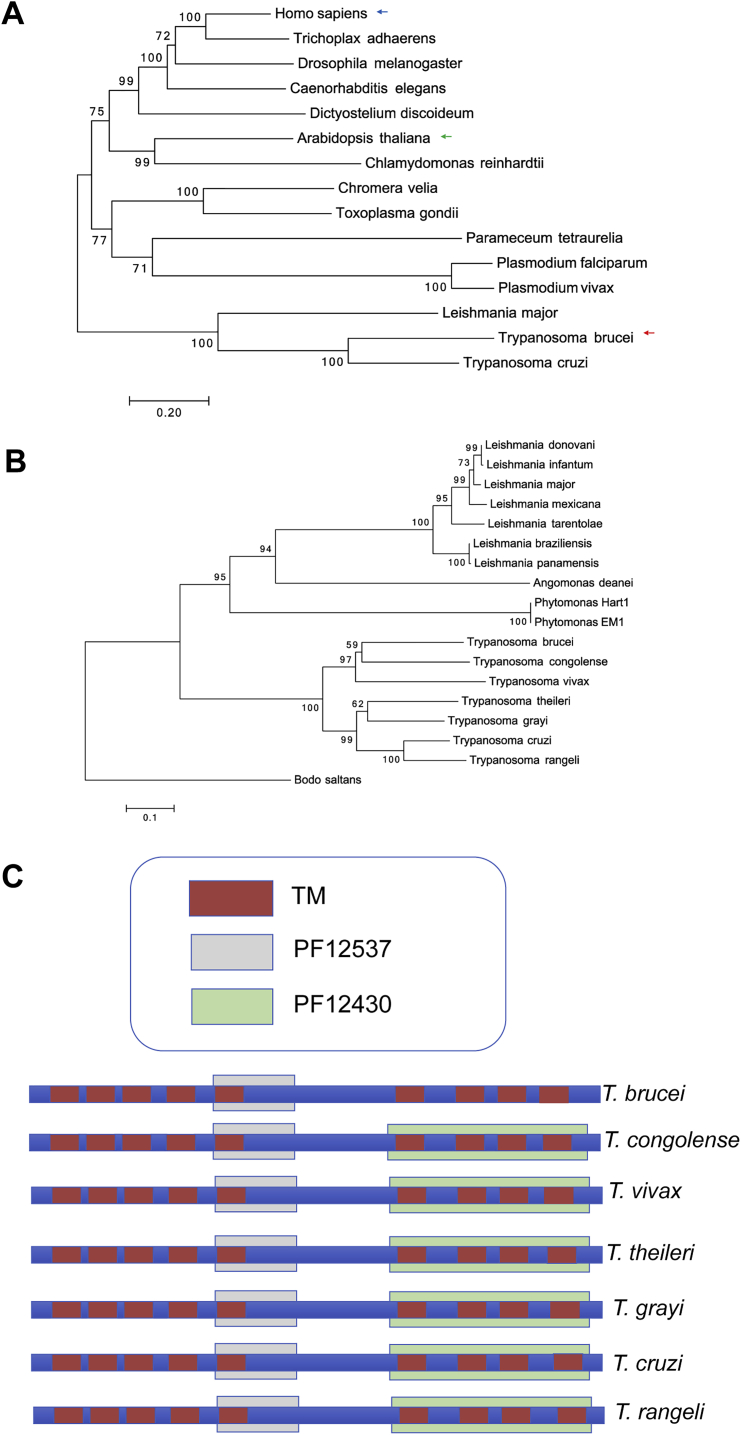
Figure S1.
GPR89 Family Members in Kinetoplastid Organisms, Related to Figure 1
(A) Phylogenetic tree of GPR89 family representatives in eukaryota. Human GPR89, Arabidopsis GTG1/GTG2 and Trypanosoma GPR89 are highlighted. The optimal tree with the sum of branch length = 7.35 is shown. The analysis involved 15 amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 383 positions in the final dataset. Accession numbers for each species are; H. sapiens, NP_001091081; T. adhaerens, XM_002112150; D. melanogaster; NP_611016; C. elegans,NP_499588; D.discoideum, XM_633754; A thaliana GTG1, NP_001031235; C. reinhardtii, XM_001695842; C. velia, Cvel_25352; T. gondii, TGME49_286490; P. tartaurelia,XM_001426347; P. falciparum, PF3D7_1008500; P. vivax, PV_094620 ; L. major, LmjF_07.0330.
(B) Phylogenetic tree of GPR89 family representatives in the kinetoplastids. The optimal tree with the sum of branch length = 4.48 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1981). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 18 amino acid sequences. All positions containing gaps and missing data were eliminated. There are a total of 302 positions in the final dataset. The tree is shown rooted on the Bodo saltans GPR89 sequence. B. saltans is a free-living nonparasitic marine kinetoplastid of the bodonid clade from which trypanosomatids descended (Jackson et al., 2016).
(C) Domain structure of GPR89 members in the kinetoplastida highlighting the position of predicted transmembrane domains (red) Pfam domain 12537 (gray) and Pfam domain 12430 (green).