Figure 6.
p53 and Myc Co-regulate Mitotic Genes in CSCs
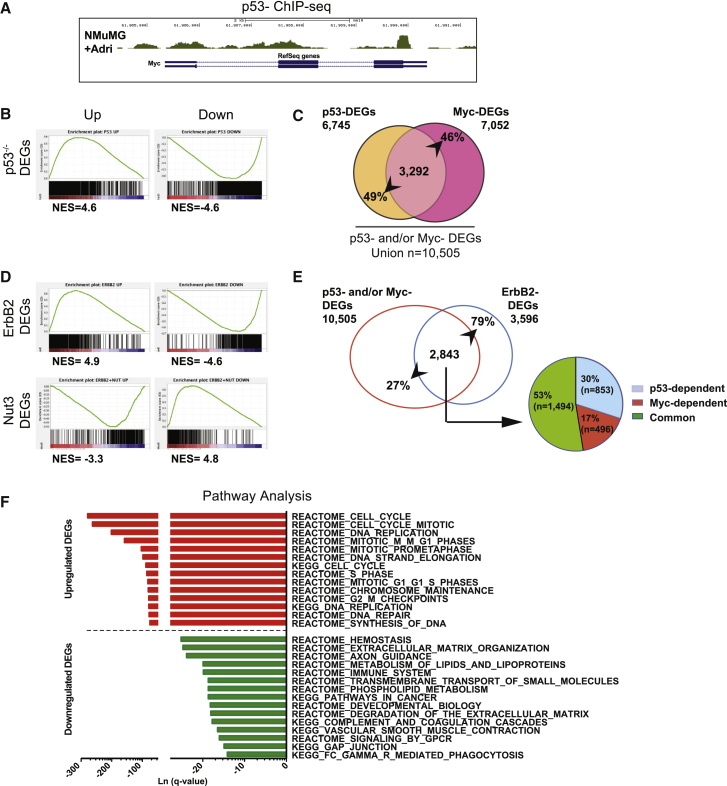
(A) Genome browser visualization of p53 binding on the c-myc gene.
(B) GSEA pre-ranked analysis showing enrichment of the Myc signature in the p53−/−. Left: upregulated genes; Right: downregulated genes. NES, normalized enrichment scored.
(C) Venn diagram of p53-Myc DEGs in non-transformed mammospheres: intercross between p53-DEGs and Myc-DEGs. Numbers represent genes coherently up- and downregulated and relative percentages.
(D) GSEA pre-ranked analysis, as in (B) showing enrichment of the Myc signature in the ErbB2-DEGs (top) and the Nut3-DEGs (bottom).
(E) Left, Venn diagram of p53 and/or Myc DEGs in ErbB2 tumors: intercross between the union of p53- and Myc-DEGs (red circle) and ErbB2-DEGs (blue circle). Right: pie chart, characterization of the p53 and/or Myc DEGs in only p53-dependent (blue), only Myc-dependent (red), and common p53-Myc DEGs (green).
(F) Pathway analysis of the ErbB2 p53-Myc common DEGs using MSigDB (http://software.broadinstitute.org/gsea/msigdb/index.jsp). The top 15 up- and downregulated pathways are shown ranked by q-value.