Figure 7.
MitSig Expression Correlates with Breast Cancer Aggressiveness and Poor Prognosis (GEO: GSE1456, GSE2034, GSE4922, GSE7390)
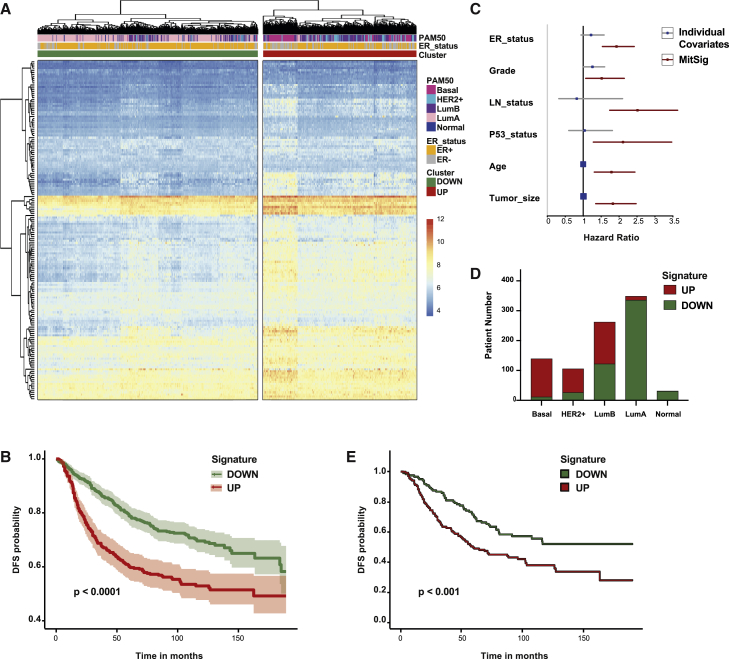
(A) Hierarchical clustering of 892 breast cancer patients according to the log2-normalized probe intensity of 161 of the 189 genes of the MitSig (values are expressed as Z score). Color code of ER status, PAM50 classification, and probe-intensity scale defined in the legend on the right.
(B and E) Disease-free survival (DFS) curve of all patients (B) and luminal B patients (E) grouped in DOWN and UP cohorts based on the expression of 161 genes of the MitSig. Log-rank test, p = 3.34e−08 and Hazard ratio (HR) = 1.93, 95% CI, 1.54–2.42 (B). Log-rank test p = 9.88e−4 and HR = 1.78, 95% CI, 1.26–2.52 (E).
(C) Forest plot for Cox multiple regression analysis of MitSig (in red) versus individual covariates (in blue). LN_status, lymph node status.
(D) Histogram showing the distribution of breast cancer patients with MitSig classification UP and DOWN within each molecular subtype (basal; HER2+; LumB, luminal B; LumA, luminal A; and normal, normal-like).