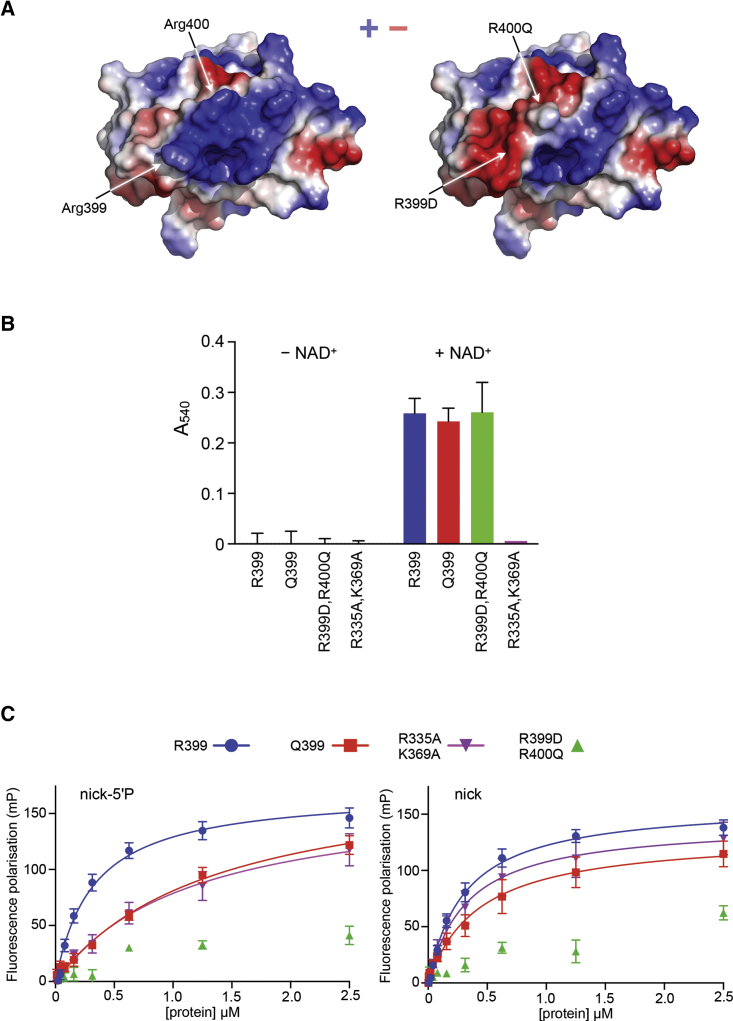
Figure 3.
Mutational Analysis of the DNA-Binding Site
(A) Solvent-accessible surface of the DNA-binding site colored by electrostatic potential (calculated in PyMol). Residues perturbed by DNA binding (including Arg399 and Arg400) map to an intensely positively charged surface patch (left), whose polarity is predicted to be reversed by the combination of R399D and R400Q mutations (right).
(B) PAR-binding assay (see STAR Methods) of XRCC1-BRCT1 variants and mutants. Both codon 399 variants and the putative DNA binding disruptive R399D,R400Q double mutant bind tightly to PAR chains generated on plates coated with histone H1 and incubated with PARP1 and NAD+, whereas no binding is seen with the R335A,K369A double mutant, which affects two residues in the PAR-binding site (Breslin et al., 2015). No binding is seen for any of the constructs in the absence of NAD+. Data represent the mean of four measurements of three separate replicates analyzed by two-way ANOVA. Error bars show ± 1 SEM.
(C) Fluorescence polarization assays of XRCC1-BRCT1 variants and mutants to FITC-labeled nicked dsDNA oligonucleotides with (left) or without (right) 5′ phosphorylation at the nick site. The codon 399 variants and the PAR-binding site mutant all bind with high affinity to both nicked duplex oligonucleotides, whereas the R399D,R400Q double mutant shows very low fluorescence poloarization (FP) values, which cannot be fitted to a binding curve (for Kd values, see Figure S1B). Data represent the mean of four measurements comprised of two separate replicates with XRCC1-BRCT1 from two separate protein purifications. Error bars show ± 1 SEM.