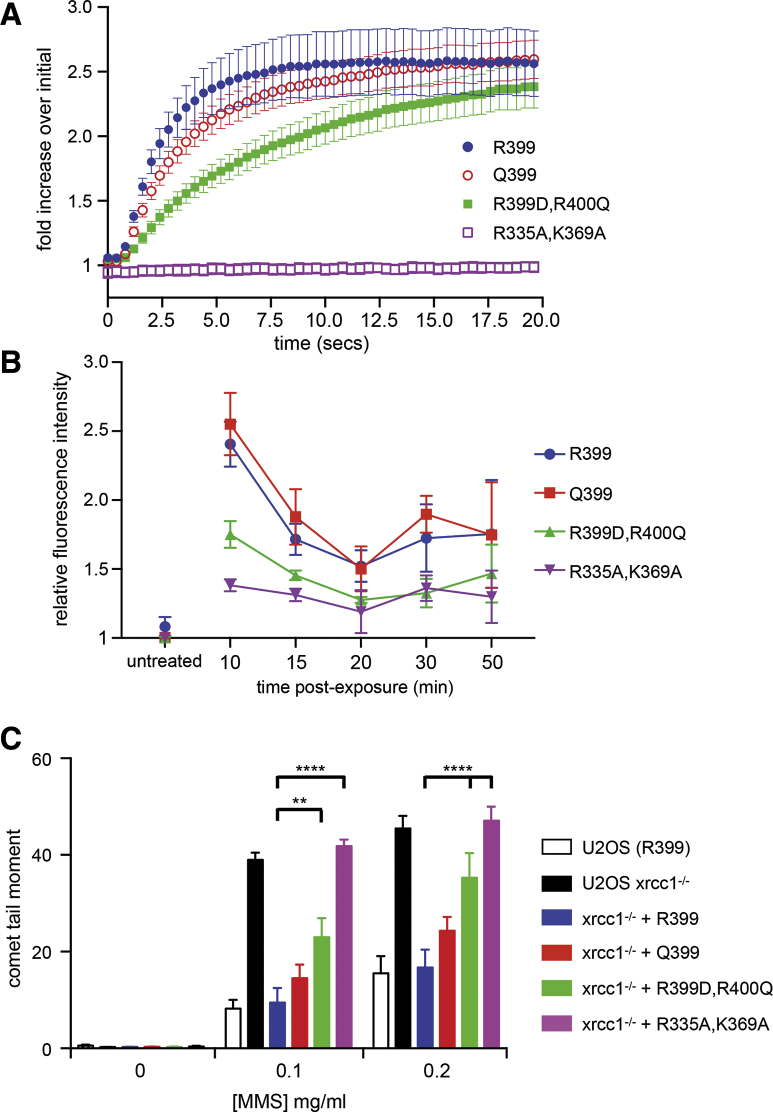
Figure 4.
DNA Binding Contributes to XRCC1-Dependent DNA Damage Repair
(A) Recruitment of XRCC1 variants and mutants to laser micro-irradiation DNA damage. Both codon 399 variants are rapidly recruited to sites of DNA damage in U2OS cells transiently transfected with GFP-XRCC1 and accumulate to comparable levels over 15–20 s post-laser exposure. Consistent with previous studies (Breslin et al., 2015), mutational disruption of XRCC1 PAR binding (R335A,K369A) abolishes XRCC1 recruitment to DNA damage in this time frame. The R399D,R400Q mutant, which is fully competent for PAR binding but defective for DNA binding in vitro, still accumulates at sites of damage but with markedly slower kinetics than the DNA-binding and PAR-binding competent constructs. Error bars are SEM for 30 cells analyzed for each curve, except for the R399D,R400Q mutant, where only 10 cells were analyzed.
(B) Retention of XRCC1 at DNA damage. Both codon 399 variants showed high levels of retention on chromatin in U2OS cells stably transfected with GFP-XRCC1 10–20 min after exposure to DNA damage by hydrogen peroxide, whereas the PAR-binding defective mutant shows much lower levels. The DNA-binding defective mutant is retained at higher levels than the PAR-binding defective mutant but markedly reduced in comparison to the unmutated variants. Data represent the mean of three measurements, with >8000 cells per sample per experiment using Perkin-Elmer Operetta software and analysed by two-way ANOVA. Error bars show ± 1 SEM.
(C) Untransformed U2OS cells, which carry the R399 XRCC1 variant, display moderate dose-dependent alkaline comet tail moments (see STAR Methods) after treatment with methyl methanesulfonate (MMS), indicative of unrepaired single-strand breaks (SSBs). U2OS cells where the XRCC1 gene is disrupted by CRISPR/Cas9 gene editing and consequently expresses undetectable levels of XRCC1 protein (Figure S2) show significantly larger tail moments indicative of much higher levels of SSBs. This repair defect can be substantially rescued by expression of GFP-XRCC1 with either codon 399 variant, but not by GFP-XRCC1 with the PAR-binding defect. Consistent with its much reduced DNA binding in vitro, its slower recruitment to laser damage, and its poorer chromatin retention post-damage, the R399D,R400Q mutant is significantly less able to rescue SSB repair in the xrcc1−/− cells. Error bars indicate SEM over three replicates (Figure S3). Average tail moments from 100 cells/sample were measured using Comet Assay IV software (Perceptive Instruments, UK) and were scored blind. Data are the average of three independent experiments. Error bars show ± 1 SEM.