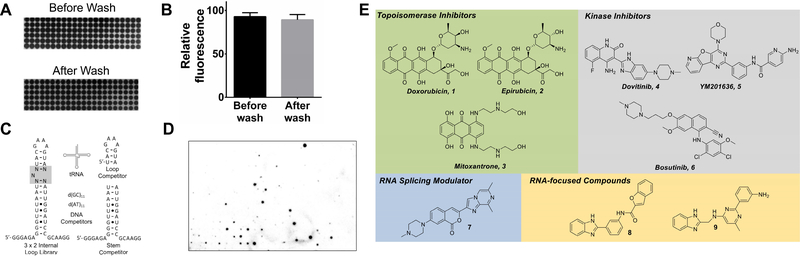
Figure 1:
The development of AbsorbArray and the RNA motif libraries used to study small molecule binding. A, representative image of an AbsorbArray chip after spotting mitoxantrone before and after washing. B, plot of the signal from mitoxantrone before and after washing. C, secondary structures of the nucleic acids used in this study. D, representative image of an AbsorbArray-enabled 2DCS screen (85.5 × 127.8 × 1.1 mm (width x height x thickness)). E, structures of various small molecules that are found to bind RNA in this AbsorbArray-enabled 2DCS experiment. See also Figure S1 and Table S1.