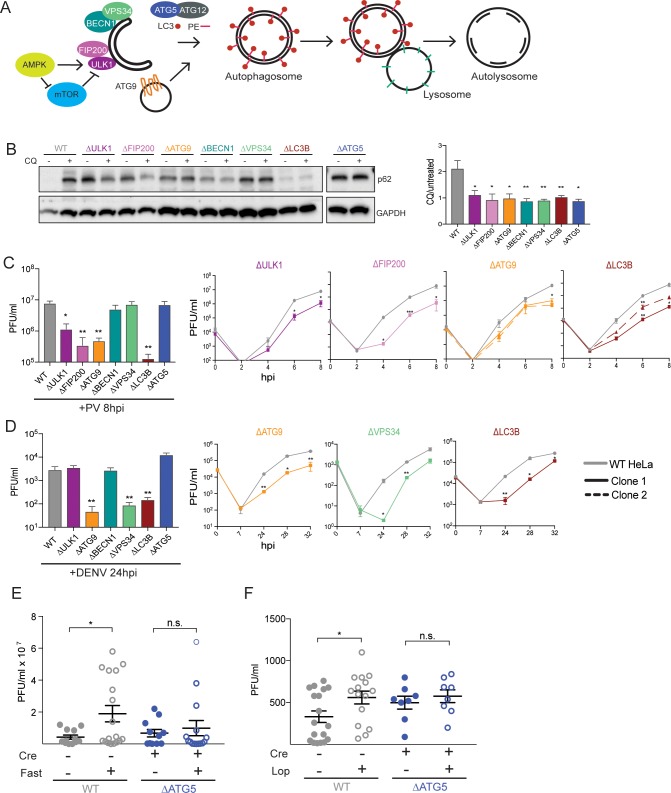
Fig 1. Diverse viruses utilize different components of the autophagy pathway.
(A) Schematic of the autophagy pathway and the components targeted in this study for KO by CRISPR-Cas9. (B) Human HeLa cells were left untreated or treated with CQ for 2 hours. Protein lysates were collected and immunoblotted for p62 and GAPDH. Quantification of autophagic flux was done by comparing p62/GAPDH in CQ and untreated samples. (C and D) HeLa cells were infected with PV or DENV at a MOI of 0.1; PFU/cell and cells (PV) or supernatant (DENV) were harvested at indicated times post infection for titration by plaque assay. (E and F) C57BL/6 PVR+/+ Atg5fl/fl Cre+/− mice were fasted for 48 hours and then infected intramuscularly with PV for E or administered loperamide every 12 hours by i.p. injection during the 4 days of infection for F. Calf muscle tissue was collected and titered by plaque assay at 4 dpi. Open circles denote fasted or drug-treated mice and blue circles indicate Atg5-deficient mice. All data are represented as mean +/− SEM. *Indicates significant P value of <0.05, **P value < 0.01, ***P value < 0.001, ****P value > 0.0001 by an unpaired t test (C and D) or a Mann–Whitney test (E–F). See also S1 and S2 Figs, S1 and S2 Tables, and S1 Data. Atg5, autophagy-related gene 5; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; CQ, chloroquine; DENV, dengue virus; dpi; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HeLa, human epithelial-derived cell line; i.p., intraperitoneal; KO, knockout; MOI, multiplicity of infection; PFU, plaque-forming units; PV, poliovirus; PVR, poliovirus receptor.