Figure 1. Ova is a niche factor for GSCs and ovary development in Drosophila.
(a) Phase contrast images of dissected ovaries from flies of indicated genotypes. Scale bar, 500 μm. (b) A graph shows the total offspring number of indicated females (n = 20, 20, 16, 16, 20 respectively). (c) Representative image of germaria from indicated genotypes labeled by α-Spectrin (red), Vasa (green), and DAPI (blue). ova1/1 and ova1/4 ovaries have numerous spherical-shaped spectrosome-containing cells (tumorous) or are empty of germline cells (germless), indicated by lack of germline cell marker Vasa (green). A wild-type (WT) germarium is usually 2 GSCs localized to the anterior tip. Scale bar, 10 μm. (d) A graph shows the percentage of normal, germless, and tumorous germaria of indicated genotypes (n = 14, 20, 30, 117, 57 respectively). (e) c587ts > ova RNAi germarium accumulated GSC-like cells after shift to 29°C for 7 and 14 days. Scale bars, 10 μm. (f) Escort cell-specific expression of ova rescued oogenesis and GSC differentiation defect of ova1/4 females. Red, α-Spectrin; Green, Vasa (g) Quantification of GSC-like cell number in germaria of indicated genotypes (n = 25, 20, 30, 117, 57, 30, 21, 31, 23 respectively). (h) Confocal sections of germaria stained by indicated antibodies or reporter. Scale bars, 10 µm. (i) Quantitative results of pMad and Dad-lacZ positive cell numbers from germaria of indicated genotypes. Values are mean ± SEM.; n > 20. P values by two-tailed Student t-test.
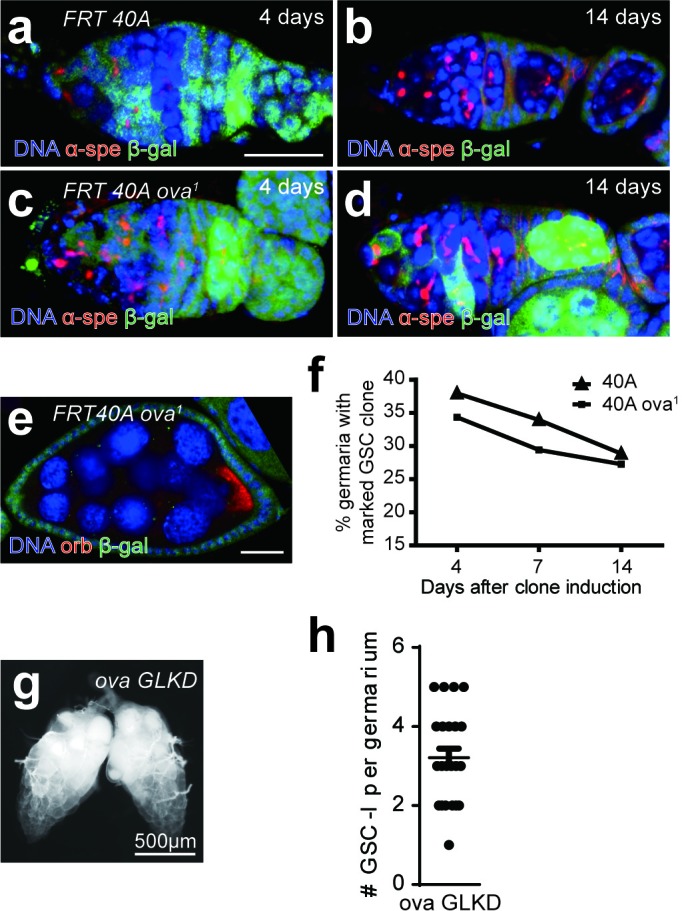
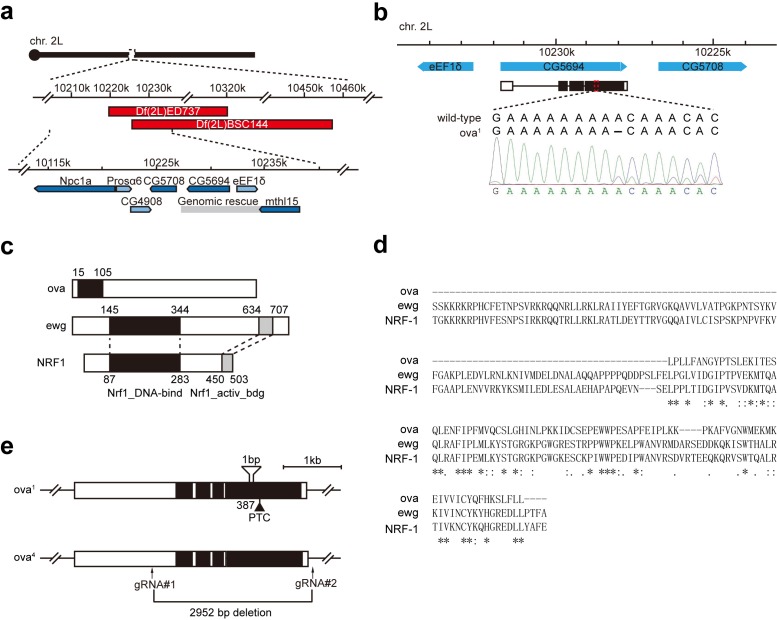
Figure 1—figure supplement 1. Ova is allelic to CG5694.
Figure 1—figure supplement 2. Ova is not cell- autonomously required for GSC differentiation.