Figure 1.
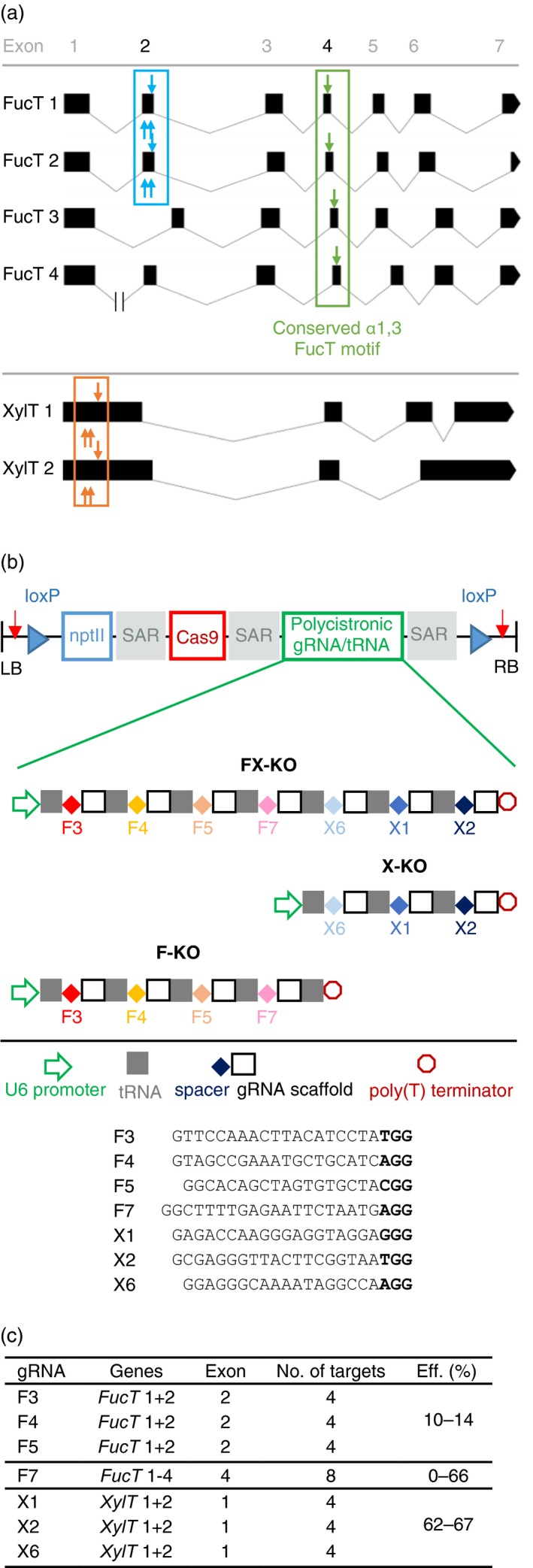
FucT 1‐4 and XylT 1 + 2 gene structure, schematic of transformation constructs and gRNA properties. (a) Structure of the four FucT genes targeted by four gRNAs and the two XylT genes targeted by three gRNAs (http://wormweb.org/exonintron). The FucT genes have seven exons, and conserved intron borders. FucT 2 has a premature stop codon in exon 7 resulting in a loss of 41 amino acids from the translated protein. FucT 4 has an unusually long intron 1 of ~7.8 kb. Three gRNAs (indicated by blue arrows) targeted exon 2 of FucT 1 and 2 (blue box), and one gRNA (green arrow) targeted the conserved catalytic motif in exon 4 of FucT 1, 2, 3 and 4 (green box). The three XylT‐specific gRNAs target exon 1 of XylT 1 and 2 (orange box). Altogether, the coloured boxes indicate the eight exons targeted by the selected gRNAs, resulting in a total of 16 targeted exons when both alleles of the six genes are considered. (b) The knockout constructs were flanked by left and right T‐DNA borders (LB and RB). To facilitate the removal of the construct from the plant genome, loxP sites (blue triangles) and human gRNA target sequences (red arrows) were included. For the selection of transformed plants on kanamycin, we included a neomycin phosphotransferase gene (nptII). The plant‐codon‐optimized cas9 gene was controlled by a hybrid 35SPPDK promoter, and the corresponding polycistronic tRNA/gRNA gene (PTG) was controlled by the A. thaliana U6 promoter. The three genes were separated by scaffold attachment regions (SARs). In the three constructs F‐KO, X‐KO and FX‐KO, four, three and seven gRNAs are expressed from the PTG, respectively, and the gRNA sequences are provided including the PAM in bold. Not drawn to scale. (c) Properties of the gRNAs, their target genes and exons, the number of genomic targets, and their mutation efficiencies (Eff.) in T0 plants.
