Figure 4.
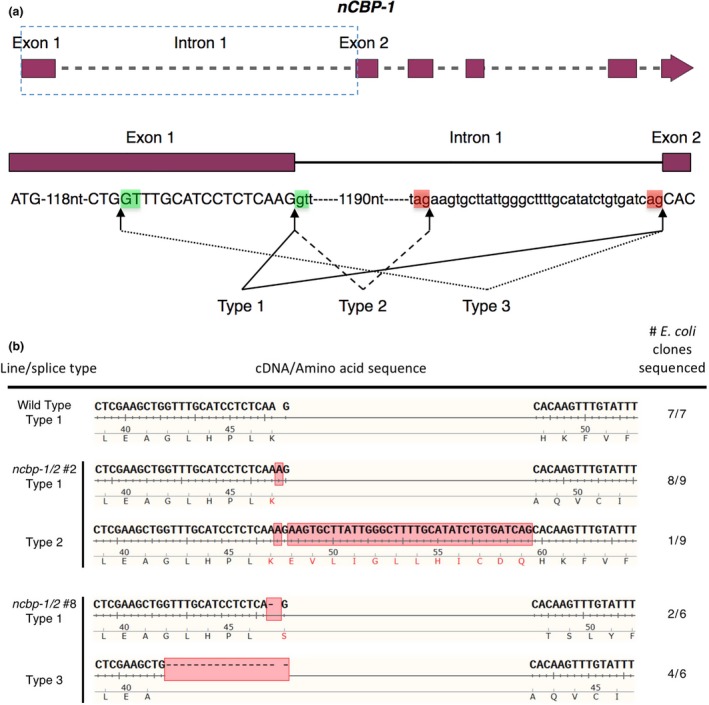
Alternative splicing of ncbp‐1 alleles is detected in ncbp‐1/2 double mutants. (a) Schematic of canonical and alternative nCBP‐1 splice sites. Boxed region of the nCBP‐1 gene model is enlarged below. Exon and intron sequences are given in capital and small letters, respectively. Green and red boxes highlight splice motifs at the 5′ and 3′ end of introns, respectively. Type 1 splicing produces the predicted wild‐type nCBP‐1 cDNA sequence. Type 2 and 3 splicing are observed in ncbp‐1/2 lines #2 & #8, respectively. (b) cDNA sequences detected in clone‐seq experiments. Red boxes denote INDELs resulting from both CRISPR/Cas9‐mediated edits and alternative splicing. In ncbp‐1/2 #2, type 2 splicing results in retention of 3′ sequence from intron 1 of ncbp‐1 (1 of 9 clones sequenced). In ncbp‐1/2 #8, an INDEL disrupting the canonical splice motif between exon 1 and intron 1 of ncbp‐1 results in a type 3 splice variant (four of six clones sequenced).
