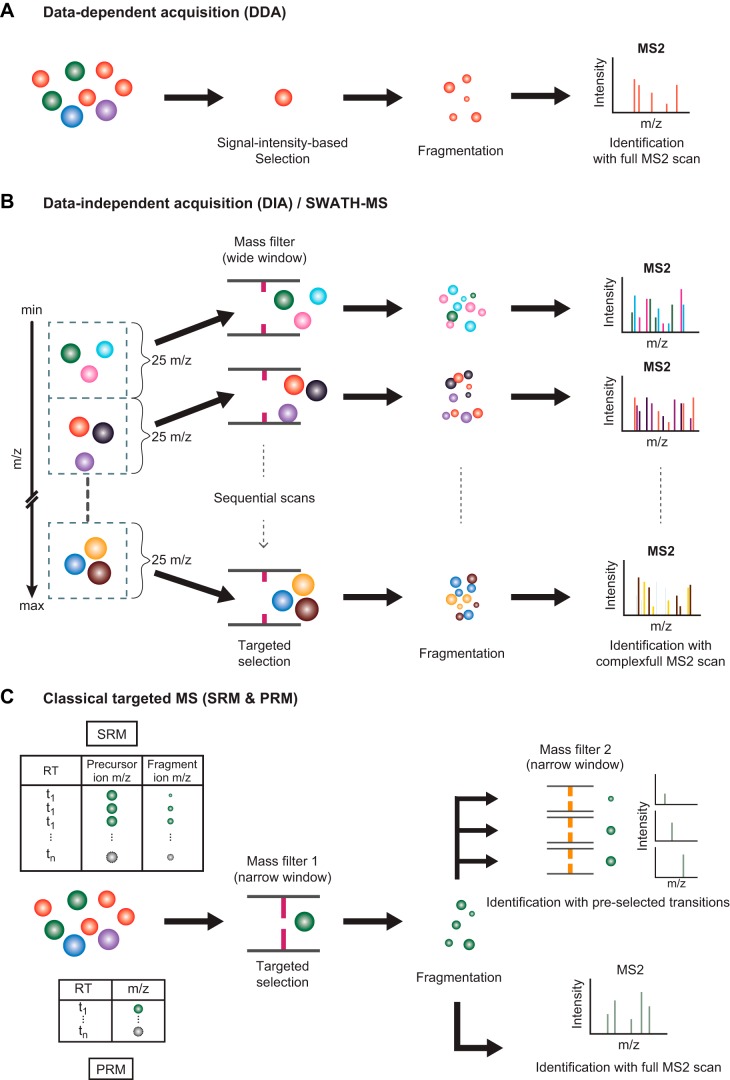
FIGURE 4.
Data acquisition techniques for mass spectrometry (MS)-based proteomics. Several modes of data acquisition are illustrated. A: data-dependent acquisition (DDA) is the most common mode for a high-throughput proteomic experiment. This mode provides a high proteome coverage, generating sequences for several thousands of peptides and identification of their cognate proteins, but has limited sensitivity for very-low-abundance peptides. B: data-independent acquisition (DIA) is an emerging technique that strives to measure every peptide in the complex samples by coisolating and fragmenting several peptides together. The resulting spectra are more complicated than DDA spectra and require specialized software for interpretation. C: selected reaction monitoring (SRM) and parallel reaction monitoring (PRM) are collectively considered as techniques for targeted proteomic study. These high-sensitivity techniques can detect lower abundance peptides but require a list of predefined peptide sequences that will be selectively isolated by the first mass spectrometer (MS1) for subsequent fragmentation and analysis by MS2 [using either pre-selected “diagnostic” fragment product ions (SRM) or a full MS2 spectrum (PRM)].