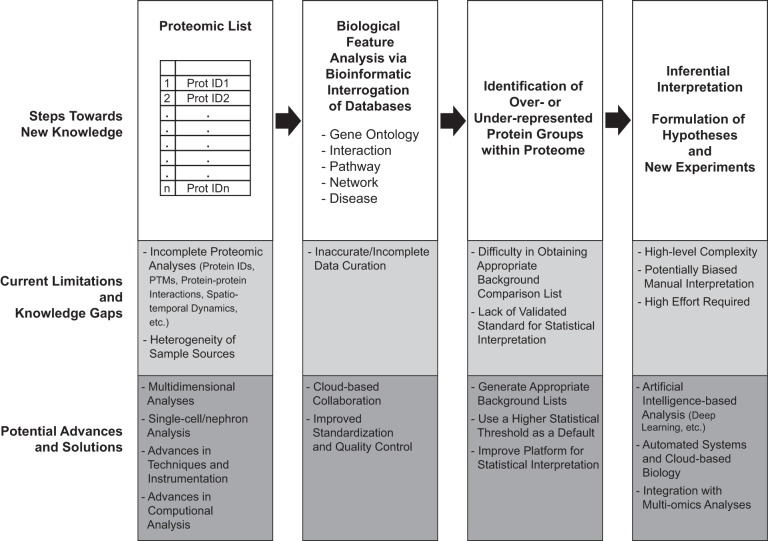
FIGURE 12.
Steps toward new knowledge, knowledge gaps, and potential advances and solutions. The creation of a proteomic list can be improved regarding both qualitative (multidimensional analyses and single-cell/tubule/nephron studies) and quantitative (fractionation techniques and instrumentation) aspects. Bioinformatic interrogation of biological databases cannot be taken as a static process, since these databases are continuously evolving and not always accurate. Thus standardization and quality control of data curation in a dynamic manner are required (191). Functional enrichment analysis is a popular method to identify over- or underrepresented protein groups. This kind of analysis is prone to several sources of bias that can confound the data interpretation, especially in the selection of background comparison list and in the statistical interpretation of high-throughput data (163). Inferential interpretation leading to the formulation of hypotheses and new experiments is a highly complex process. Machine-learning techniques, such as deep learning, have gained much attention over the last few years. Even though these techniques cannot yield an explicit hypothesis or understanding of the underlying mechanisms, they help us draw biologically relevant conclusions from the omics data (23). Lastly, new experiments to validate the previous findings and test new hypotheses are essential, the task that always requires high effort. Automated systems and cloud-based biology are undertaken as a future direction (167a).