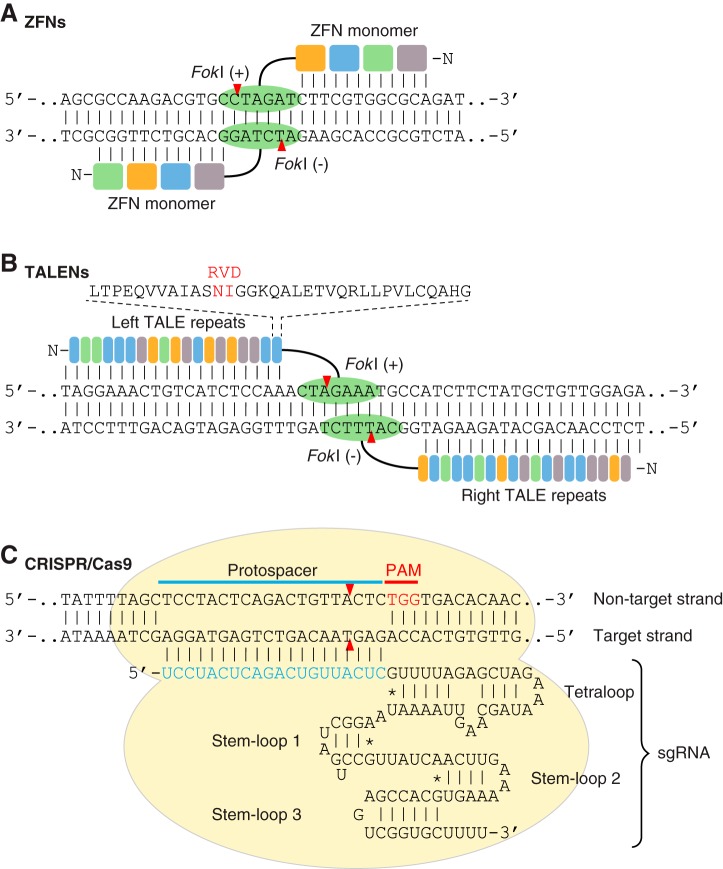
FIGURE 2.
Programmable nucleases used for genome editing. A: a schematic illustration of a pair of zinc-finger nuclease (ZFN) monomers bound to DNA. ZFN is a chimeric endonuclease composed of multiple zinc finger domains (colored boxes) at the NH2 terminus for DNA binding and a Fok1 nuclease domain (green oval) at the COOH terminus for DNA cleavage. Dimerization of two Fok1 nucleases induces a DNA double-strand break (DSB) with 4 bp of 5′ overhang. B: a schematic illustration of a pair of transcription activator-like effector nucleases (TALENs) bound to DNA. TALEN is a chimeric endonuclease composed of multiple TALE repeats (colored rectangles) at the NH2 terminus and a Fok1 nuclease domain (green oval) at the COOH terminus. Each TALE repeat recognizes 1 bp of DNA, and the sequence specificity is determined by repeat-variable diresidues (RVD; shown in red). TALEN-mediated DNA DSBs are induced by dimerization of two Fok1 nucleases. C: a schematic illustration of the engineered CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR associated protein 9) system from Streptococcus pyogenes. In the CRISPR/Cas9 system, target recognition is mediated by DNA hybridization with a single guide RNA (sgRNA), which is an engineered RNA chimera composed of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). CRISPR/Cas9-mediated DNA DSB requires a protospacer adjacent motif (PAM; shown in red), and cleavage is induced at the nucleotide 3 bp proximal to the PAM. Red arrowheads indicate cleavage site.