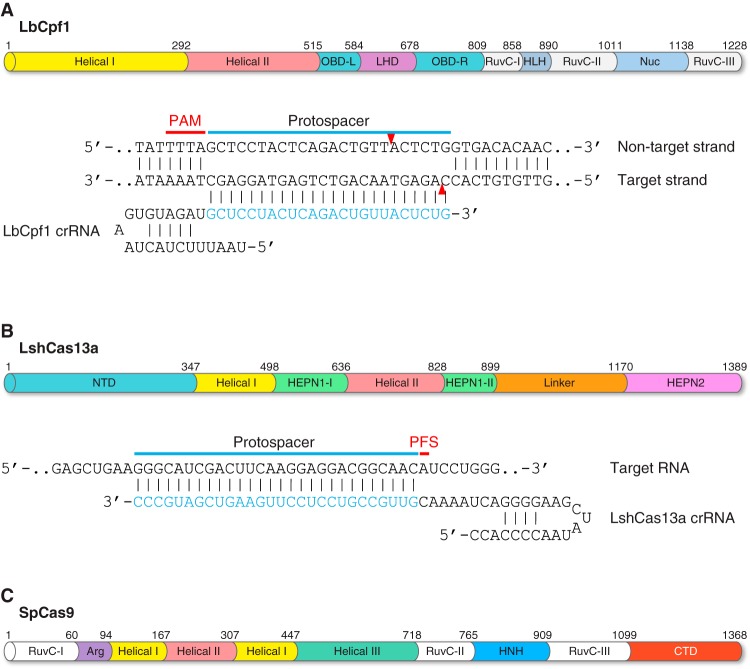
FIGURE 5.
Novel CRISPR/Cas systems. Two novel class 2 CRISPR/Cas systems have been engineered for nucleic acid recognition and cleavage, such as Cpf1 (CRISPR from Prevotella and Francisella 1) and Cas13a (formerly C2c2). A: domain organization of the LbCpf1 protein discovered in Lachnospiraceae bacterium ND2006. All Cpf1 orthologs have two nuclease domains: 1) the RuvC domain which cleaves the nontarget DNA strand and 2) the Nuc domain which cleaves the target DNA strand. The LbCpf1 crRNA is shown hybridizing with its DNA target. The PAM is highlighted in red. Red arrowheads indicate cleavage site. B: domain organization of the LshCas13a protein discovered in Leptotrichia shahii. Cas13a has dual RNase activities, one specific for pre-crRNA processing and maturation, which is catalyzed by the helical-I domain, and the other one for RNA-guided single-stranded RNA (ssRNA) degradation, which is catalyzed by the HEPN1 and HEPN2 domains. The LshCas13a crRNA is shown hybridizing with its RNA target. CRISPR/Cas13a-mediated ssRNA cleavage is independent of a PAM; instead, it requires a 3′-protospacer flanking site (PFS; shown in red). C: domain organization of the SpCas9 protein discovered in Streptococcus pyogenes. The RuvC nuclease domain cuts the nontarget strand. The HNH nuclease domain cuts the target strand that hybridizes with the sgRNA. CTD, COOH-terminal domain.