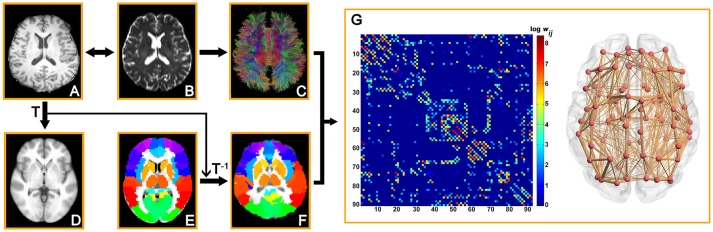
Figure 1.
Flowchart for construction of the WM structural network by DTI. (1) Coregistration from an individual T1-weighted image (A) to a DTI b0 image (B). (2) Nonlinear registration from the T1-weighted image in the native DTI space to the ICBM152 T1 template in the MNI space (D). (3) Application of the inverse transformation (T−1) to the AAL atlas in the MNI space (E), which results in individual-specific parcellation in the native DTI space (F). (4) The reconstruction of the whole-brain WM fibers (C) was performed using deterministic tractography in Diffusion Toolkit. (5) The weighted networks of each subject (G) were created by computing the number of the streamlines that connected each pair of brain regions. The connection matrix and 3D representation (axial view) of the WM structural network of a representative healthy subject are shown in the right panel. The nodes are located according to their centroid stereotaxic coordinates and the edges are sized according to their connection weights.