Figure 5.
RNA Sequencing of HCM and Isogenic Control iCMs
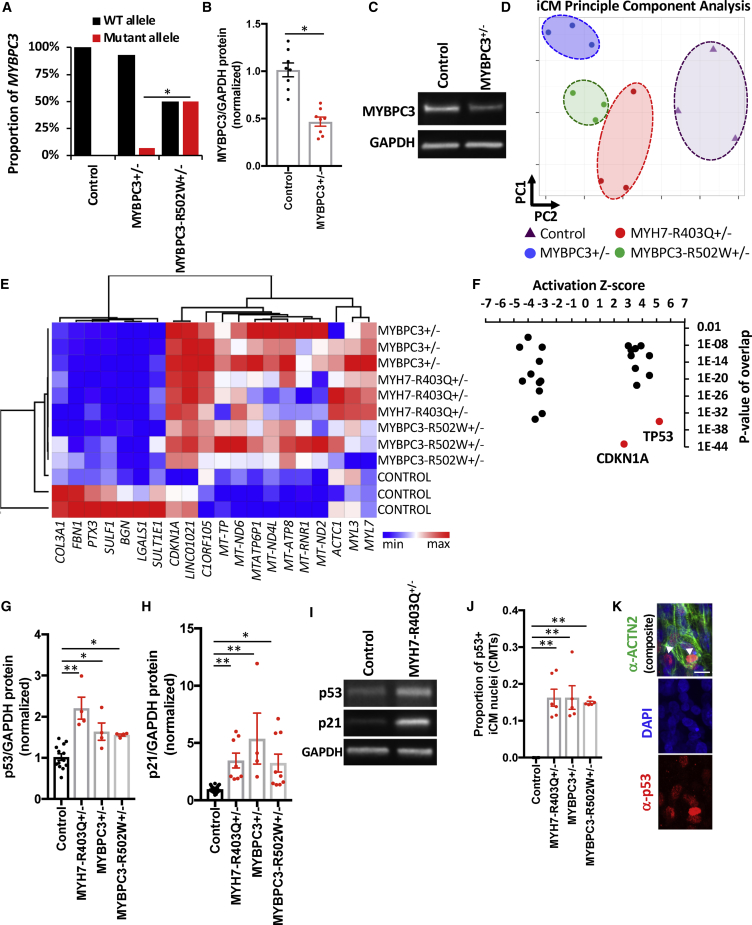
(A) By allele-specific analysis of gene transcripts obtained from isogenic control, MYBPC3+/−, and MYBPC3-R502W+/– iCMs; MYBPC3 transcripts were quantified for control (wild-type [WT]; black bar) and mutant (red bar) alleles and shown as the proportion of total MYBPC3 expression.
(B) Densitometry of immunoblots from protein lysates derived from control and MYBPC3+/− iCMs, and probed for MYBPC3 (note: truncated MYBPC3 was not identified) and for protein loading with GAPDH.
(C) Representative immunoblot from (B).
(D) Principal-component analysis (PCA) of RNA transcripts from three biological replicates of isogenic control (purple triangles) and MYBPC3+/− (blue circles), MYBPC3-R502W+/– (green circles), and MYH7-R403Q+/– iCMs (red circles).
(E) Hierarchical clustering of genes contributing to PC1 and PC2 from (D) and illustrated by heatmap (Table S2).
(F) Differentially expressed gene transcripts (log2FC > 0.3 or < –0.3 and false discovery rate < 0.1) were analyzed by pathway analysis using Ingenuity Pathway Analysis and identified pathways (black and red dots) were organized by activation Z score and p value of overlap.
(G–I) Densitometry of immunoblots from control and HCM iCM protein lysates, normalized for protein loading (GAPDH) (G) and probed with antibodies to p53 or (H) p21. See representative blots in (I).
(J and K) Quantification of iCM p53+ nuclei from confocal images of fixed CMTs immunostained with an antibody to p53 (red), cardiomyocyte-specific ACTN2, and DAPI co-stain (J). See representative image in (K), arrowhead marks p53+ nuclei that co-stain with ACTN2. Scale bar, 15 μm.
Significance (∗p < 0.05 and ∗∗p < 0.001) was assessed by Fisher's exact test (A), Student's t test (B), or ANOVA (G, H, and J); and data are means ± SEM (error bars) (B, G, H, and J). Each data point represents a sample generated from a batch of iCMs (B, D, E, G, and H) or single CMT (J) generated by at least three biological replicates by iPS differentiation batch.