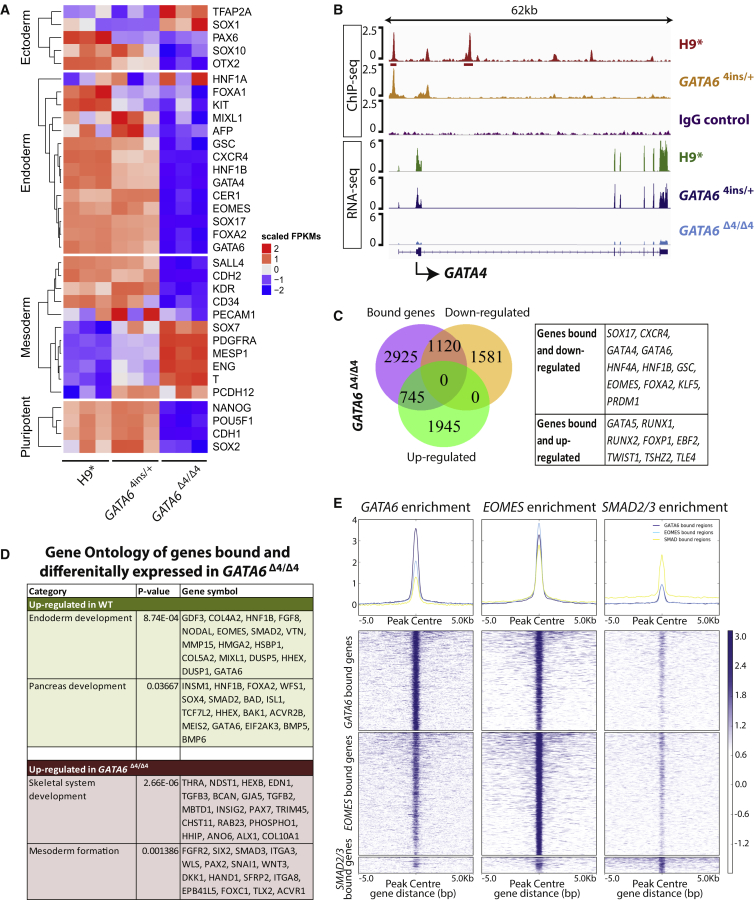
Figure 3.
GATA6 Is a Key Regulator of the DE Transcriptional Network
(A) Heatmap illustrating differential gene expression of key germ layer markers via RNA-seq between H9∗ cells and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at the DE stage. n = 3 biological replicates for each cell line.
(B) ChIP-seq binding profiles of H9∗ and GATA64ins/+ showing GATA6 enrichment near GATA4, and GATA4 expression by RNA-seq in H9∗ and H9-derived GATA64ins/+ and GATA6Δ4/Δ4 mutant cells at the DE stage. The input control profile (IgG control) is included for comparison. The ChIP-seq binding profile is derived from merging two biological replicates.
(C) Venn diagram indicating the overlap of GATA6-bound genes from ChIP-seq at the DE stage with downregulated or upregulated genes in H9-derived GATA6Δ4/Δ4 mutant cells compared with H9∗ cells by RNA-seq. Key bound genes up- or downregulated are indicated in the table.
(D) Enriched gene ontology showing developmental pathways from direct target genes differentially expressed between H9∗ and H9-derived GATA6Δ4/Δ4 mutant cells derived from BETA analysis.
(E) Density heatmaps of GATA6-binding peak intensity at DE indicating direct overlap with known endodermal regulators, including SMAD2/3 and EOMES, within a 5-kb window centered at the transcription start site.