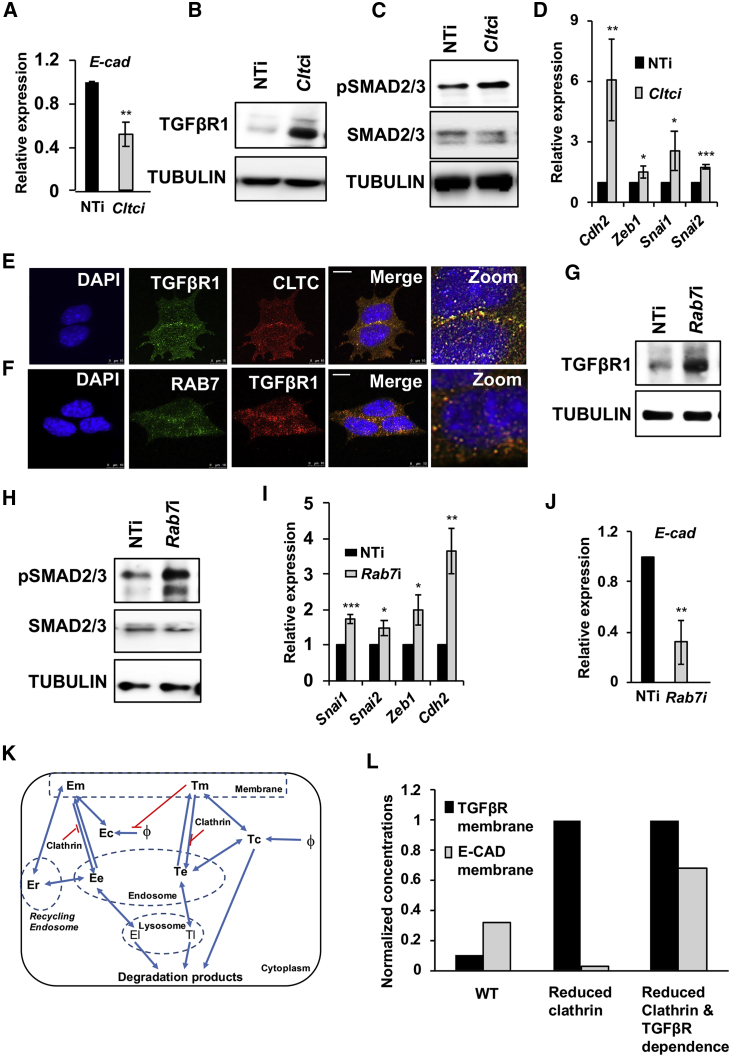
Figure 3.
CME Promotes Degradation of TGF-βR1 to Maintain E-CAD Levels in mESCs
(A) Graph showing levels of the E-cad transcript by qRT-PCR analysis upon knockdown of Cltc in mESCs. NTi, non-targeting siRNA control; Cltci, Cltc siRNA.
(B and C) Western blots showing TGF-βR1 (B), pSMAD2/3, and total SMAD2/3 (C) levels in mESCs, 72 hr post Cltc knockdown.
(D) Graph showing levels of indicated genes by qRT-PCR analysis after 3 days of Cltc knockdown in mESCs.
(E and F) Representative micrographs showing the co-localization between CLTC and TGF-βR1 (E) and between TGF-βR1 and RAB7 in mESCs (F). Scale bar, 10 μm.
(G and H) Western blots showing TGF-βR1, pSMAD2/3, and total SMAD2/3 levels 72 hr post Rab7 knockdown in mESCs. Rab7i, Rab7 siRNA.
(I) Graph showing levels of indicated genes by qRT-PCR analysis upon knockdown of Rab7 in mESCs.
(J) Graph showing levels of E-cad by qRT-PCR analysis upon knockdown of Rab7 in mESCs.
(K) Construction of a model to show the inter-relationships between TGF-βR, E-CAD, and CLTC in mESCs. E, E-CAD; T, TGF-βR; m, membrane; c, cytoplasm; e, endosome; r, recycling endosome; l, lysosome.
(L) Graph showing levels of E-CAD and TGF-βR based on simulation of the constructed model. WT (wild-type) represents the pre-treatment concentrations of membrane-bound TGF-βR and E-CAD. The “reduced clathrin” set shows the steady-state values following clathrin depletion. The “reduced clathrin and TGF-βR dependence” set is obtained by simulating a decrease in TGF-β signaling following CLTC depletion, by changing the parameter Kitmec, which changes the sensitivity of the E-CAD formation rate to TGF-βR levels by 5-fold, keeping other parameters constant.
Error bars represent mean ± SD from three independent experiments (n = 3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 by two-tailed Student’s t test.