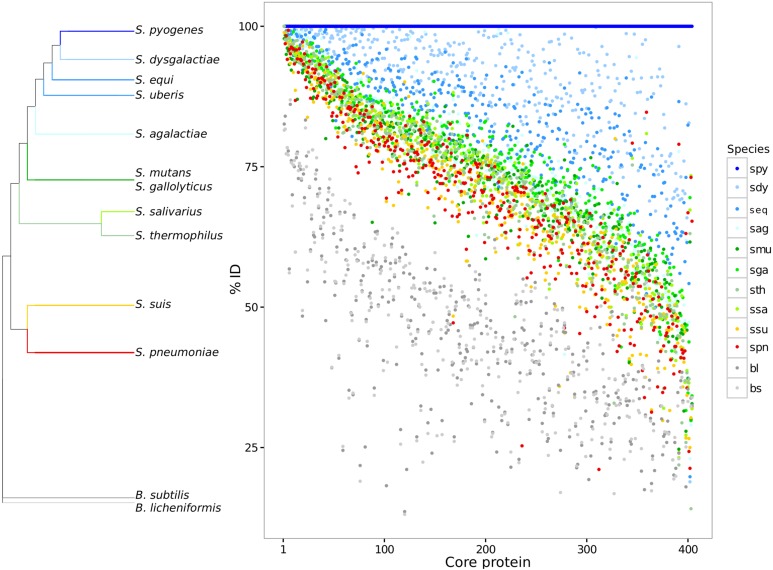
Figure 1. Core genome variability amongst different streptococci clades.
Each core protein, for each streptococci species, was aligned against the reference S. pyogenes. The pairwise identity of each core protein, calculated by global sequence alignment, was sorted and plotted. The dendrogram shows a summary of genomic similarity score (GSS) distances. The identity variability highlights the species diversity even for the conserved coding genes. S. pyogenes (spy), S. dysgalactiae (sdy), S. agalactiae(sag), S. parauberis (spu), S. iniae (sin), S.uberis (sub), S. equi subsp. zooepidemicus (seq_z), S. equi ssp., equi (seq_z), S. suis (ssu), S. thermophilus (sth), S. salivarius (ssa), S. mutans (smu), S. intermedius (sint), S. oligofermentans (sol), S. sanguinis (ssan), S. gordonii (sgo), S. parasanguinis (sps), S. pasteurianus (spas), S. oralis (sor), S. pneumoniae (spn), S. pseudopneumoniae (sppn), S. mitis (smi), S. gallolyticus (sga), S. macedonicus (sma), S. lutetiensis (slu), S. infantarius (sinf), B. subtilis (bs), and B. licheniformis (bl).