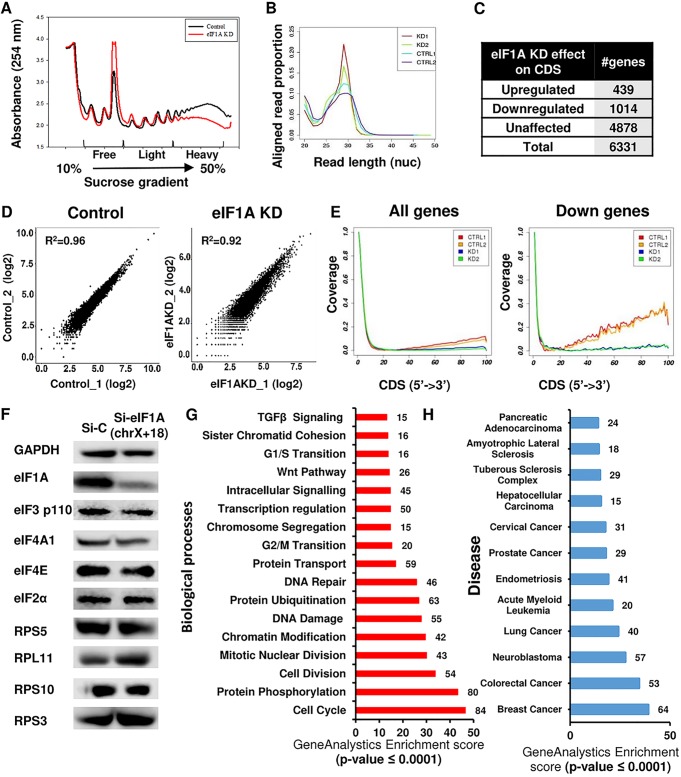
FIG 2.
Ribosomal profiling of eIF1A KD cells reveals translation downregulation of cell cycle- and cancer-associated genes. (A) eIF1A depletion dramatically affects global translation. Cell lysates of MEFs treated with control siRNA (black) or eIF1A siRNA (red) were subjected to sucrose gradient sedimentation and fraction collection to obtain polysome profiles. (B) Ribosome-protected fragment length (in nucleotides [nuc]) in control and eIF1A KD samples (n = 2 per sample category). (C) Summary of the number of genes whose ribosomal occupancy was affected (fold change, ≥2 or −2) or unaffected in coding regions (CDS) in response to eIF1A KD. (D) Scatter plot showing the reads per kilobase per million correlations (R2 > 0.9) between the 2 independent replicate experiments with control and eIF1A KD MEFs. (E) Meta-gene analysis of the distribution of normalized reads in the coding region (CDS) of all analyzed genes (n = 6,331) and the downregulated genes (n = 1,014) (down). (F) Control and eIF1A KD cells were analyzed by Western blotting with antibodies against eIF1A, GAPDH, and the indicated eIFs and ribosomal proteins. chrX+18, chromosomes X and 18. (G) Gene enrichment analysis of the biological processes associated with the CDS downregulated genes. The number of genes in each category and the P value are indicated. TGF, tumor growth factor. (H) Gene enrichment analysis of diseases associated with the CDS downregulated gene. The number of genes in each category and the P values are indicated.