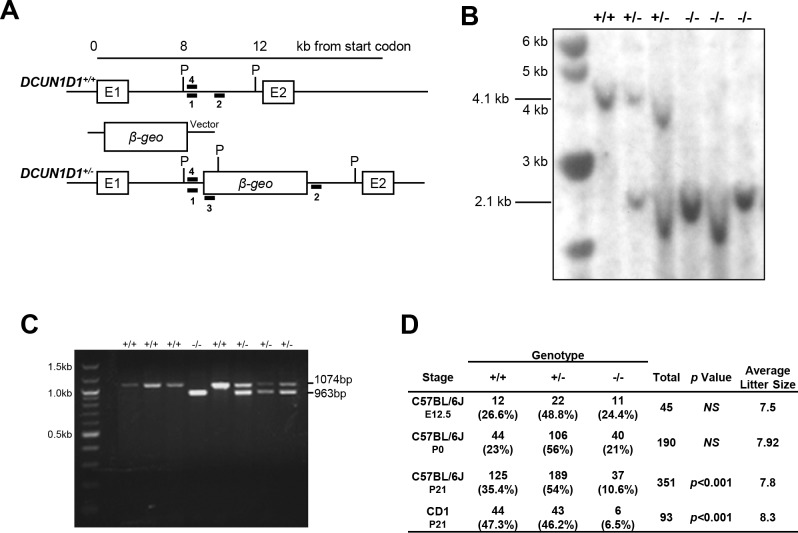
Fig 1. DCUN1D1-/- mice are viable.
(A) Schematic diagram of the mouse DCUN1D1 gene locus (top), targeting vector (middle, pGT0lxf), and the disrupted mutant allele following homologous recombination (bottom). The targeting vector contains the β-galactosidase coding sequence inserted between exon 1 (E1) and exon 2 (E2) of DCUN1D1. The locations of PCR genotyping 5’ forward (1), 3’ reverse (2), and vector-specific reverse (3) primers used to distinguish mutant from wild-type alleles are shown. The location of the 5’ probe (4) used for Southern blotting analysis within the PvuII digestion sites (P) is indicated. (B) Results from Southern blotting of tail DNAs from F1 progeny of heterozygote intercrosses digested with PvuII. A 2.1-kb fragment indicates heterozygote. (C) PCR analysis of DNA extracted from DCUN1D1+/+, DCUN1D1+/-, and DCUN1D1-/- mice with primers 1, 2, and 3 denoted in Fig 1A. A product of 963 bp indicates heterozygote. (D) Results from mating DCUN1D1+/- mice, showing normal litter sizes and maintenance of expected Mendelian ratios at E12.5 and birth (P0). There was a reduction in viability in DCUN1D1-/- mice on postnatal day 21 (P21) in both C57BL/6J and CD1 outbred strains (rows 3 and 4, respectively). P values are derived from χ2 analyses.