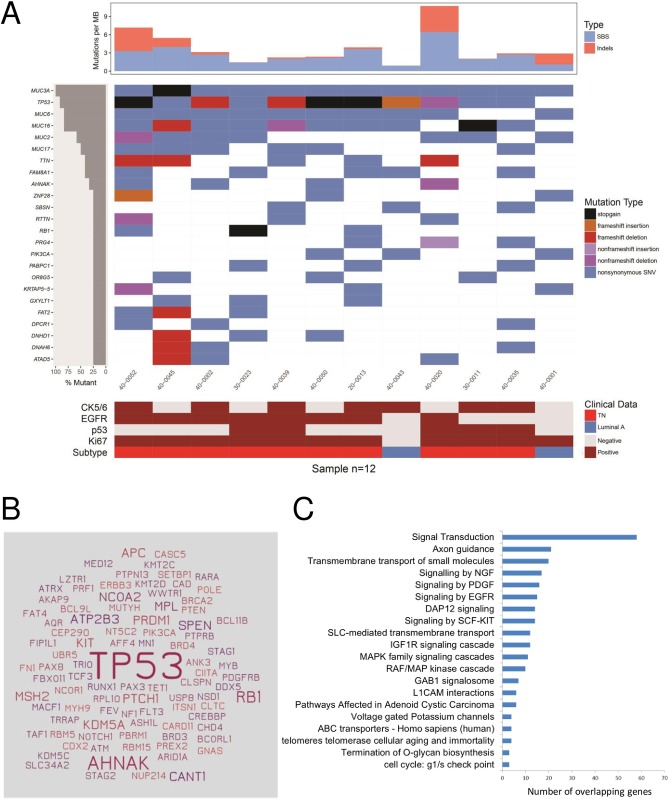
Fig 2. Whole-exome sequencing results in 12 PRECAMA samples.
Only coding non-silent somatic mutations are considered. (A) Mutation rates (top panel), top mutated genes and their mutation types (middle panel), and IHC features (lower panel), sorted by top mutated genes. Luminal A: ER+/HER2-; triple-negative: ER-/PR-/HER2-. (B) All cancer genes somatically mutated in the 10 TN samples are depicted; the size of gene names is proportional to the number of samples mutated for each gene. (C) Pathways enriched (q-value <0.05) in the list of genes mutated in TN cases with allele frequency >20% and predicted deleterious/probably deleterious by PolyPhen-2 (N = 333 genes). Number of overlapping genes in each pathway is shown.