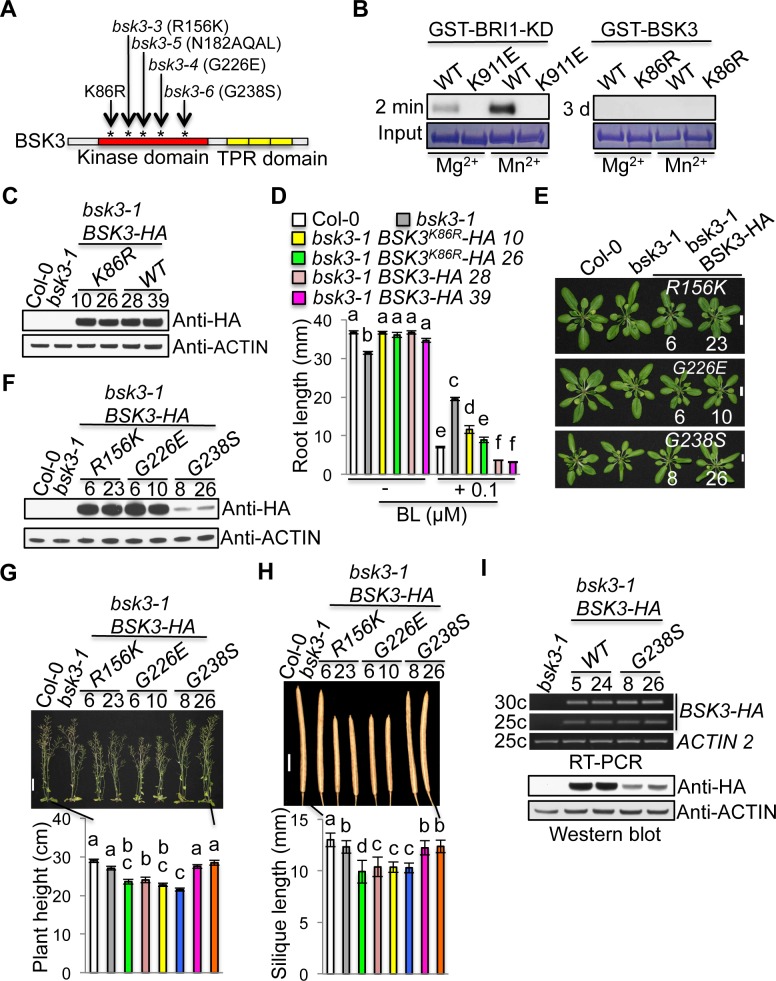
Fig 3. Kinase dead BSK3K86R protein partially rescues the bsk3-1 mutant phenotypes.
(A) BSK3 mutations in the kinase domain. The kinase domain (58–312 aa) of BSK3 was predicted by SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de/). (B) In vitro kinase assays. About 10–20 μg of GST fusion proteins were loaded for kinase assays. Dried protein gels were exposed to films for 2 minutes (GST-BRI1-KD and GST-BRI1-KDK911E) or 3 days (GST-BSK3 and GST-BSK3K86R) at—80 oC. (C) Western blot analyses of BSK3-HA and BSK3K86R-HA protein expression. (D) Primary root length of 7-day-old light-grown seedlings. Seedlings were grown on ½ LS agar plates with or without 0.1 μM BL for 7 days. Error bars represent SEM (n = 40–50). (E) Four-week-old plants. Primary bolts were removed to better observe rosette leaves. Scale bar = 1 cm. (F) Western blot analyses of BSK3R156K/G226E/G238S-HA protein expression. (G) Seven-week-old plants. Error bars represent SEM (n = 14–19). Scale bar = 4 cm. (H) Mature siliques. Siliques from 14 to 19 plants (10 siliques per plant) were analyzed. Error bars represent STD (n = 140–190). (I) Semi-quantitative RT-PCR and western blot analyses of BSK3-HA and BSK3G238S-HA transcript and protein levels, respectively. Homozygous T4 transgenic plants were analyzed. (C, F, and I) Thirty micrograms of total proteins from 7-day-old light-grown seedlings were loaded. (D, G, and H) Different letters above the bars indicate significant differences (P < 0.05).