The capacity to sense and adapt to changing carbon dioxide levels is crucial for all organisms. In fungi, CO2 is a key determinant involved in fundamental biological processes, including growth, morphology, and virulence. In the pathogenic fungus Candida albicans, high CO2 is directly sensed by adenylyl cyclase to promote hyphal growth. However, little is known about the mechanism by which hyphal development is maintained in response to physiological levels of CO2. Here we report that a signal transduction system mediated by a phosphatase-kinase pair controls CO2-responsive Ume6 phosphorylation and stability that in turn dictate hyphal elongation. Our results unravel a new regulatory mechanism of CO2 signaling in fungi.
KEYWORDS: Candida albicans, Ssn3/Cdk8, Ume6, carbon dioxide signaling, hyphal development
ABSTRACT
Candida albicans is the most common cause of invasive fungal infections in humans. Its ability to sense and adapt to changing carbon dioxide levels is crucial for its pathogenesis. Carbon dioxide promotes hyphal development. The hypha-specific transcription factor Ume6 is rapidly degraded in air, but is stable under physiological CO2 and hypoxia to sustain hyphal elongation. Here, we show that Ume6 stability is regulated by two parallel E3 ubiquitin ligases, SCFGrr1 and Ubr1, in response to CO2 and O2, respectively. To uncover the CO2 signaling pathway that regulates Ume6 stability, we performed genetic screens for mutants unable to respond to CO2 for sustained filamentation. We find that the type 2C protein phosphatase Ptc2 is specifically required for CO2-induced stabilization of Ume6 and hyphal elongation. In contrast, the cyclin-dependent kinase Ssn3 is found to be required for Ume6 phosphorylation and degradation in atmospheric CO2. Furthermore, we find that Ssn3 is dephosphorylated in 5% CO2 in a Ptc2-dependent manner, whereas deletion of PTC2 has no effect on Ssn3 phosphorylation in air. Our study uncovers the Ptc2-Ssn3 axis as a new CO2 signaling pathway that controls hyphal elongation by regulating Ume6 stability in C. albicans.
INTRODUCTION
Candida albicans is a common opportunistic fungal pathogen of humans. As a part of the commensal microbiota, C. albicans is a benign inhabitant of the gastrointestinal and genitourinary tracts most of the time. However, it can infect sites ranging from the skin and the oral and vaginal mucosa to deep tissues if host or environmental factors are permissive (1). Disseminated invasive candidiasis has an estimated mortality rate of 40%, even with the use of antifungal drugs (2). With the limited types of antifungal drugs available and rising populations of susceptible patients, there is a pressing need for understanding mechanisms of Candida pathogenesis in order to develop new approaches for treating invasive candidiasis.
Numerous traits that contribute to virulence have been documented for C. albicans, and among the most prominent is its ability to grow either as a unicellular budding yeast or in filamentous forms (3). Unlike dimorphic fungal pathogens of humans (e.g., Histoplasma capsulatum, Paracoccidioides brasiliensis, and Talaromyces [formerly Penicillium] marneffei) that normally grow in filamentous forms outside the human body but convert to yeast form in human tissues (4), C. albicans is able to switch reversibly between yeast, pseudohyphae, and hyphal growth forms, and is found in both yeast and filamentous forms in the host (5). The hyphal form plays key roles in the infection process, and has a variety of specific properties linked to virulence, including adherence (6, 7), secretion of hydrolases (8), and candidalysin (9), to damage host cells. Hypha-specific genes UME6 and HGC1 are regulators of hyphal transcription and morphogenesis (10–12). Levels of the transcription factor Ume6 control the levels and duration of hypha-specific transcription (13). The yeast-to-hypha transition requires initiation and then maintenance. Hyphal initiation requires a rise in temperature to 37°C and release from quorum sensing molecules, such as farnesol, to temporarily clear the major repressor of hyphal morphogenesis, Nrg1 (14, 15). Hyphal maintenance requires active sensing of the surrounding environment. Nutrient limitation, serum, or N-acetylglucosamine activates the expression of transcription factor Brg1 to recruit the Hda1 histone deacetylase to promoters of hypha-specific genes, leading to nucleosome repositioning, obstruction of Nrg1 binding sites, and sustained hyphal development (16). In parallel to the nutrient-responsive chromatin-remodeling pathway, the combination of hypoxia and high CO2, but neither condition alone, maintains hyphal elongation by stabilizing the transcription activator Ume6, leading to sustained hyphal development through a positive feedback loop in which Ume6 activates its own transcription (17). The Ume6 stabilization and chromatin-remodeling pathways act in parallel to control hyphal development and virulence during disseminated infection. Ofd1, a prolyl 4-hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase, acts as an oxygen sensor that regulates Ume6 stability in response to hypoxia (17, 18). Ume6 stability in C. albicans is also controlled by the level of CO2. However, the signaling pathway for the CO2-responsive Ume6 stabilization in hyphal elongation remains elusive in C. albicans.
Reversible protein phosphorylation is a key protein modification involved in the regulation of numerous physiological processes. It is an extremely common event in signal transduction, and it is considered the main mechanism of posttranslational modification leading, for instance, to a change in enzyme activity. The phosphorylation state of a protein results from the balance of protein kinases and protein phosphatase activities. Ssn3 and its cyclin Ssn8, along with the cofactors Srb8/Med12 and Srb9/Med13, form the kinase submodule of the RNA polymerase-associated mediator complex that functions as a bridge between basal transcription machinery and gene-specific transcriptional factors (19). Ssn3 has also emerged as a key regulator of multiple transcriptional programs linked to nutrient sensing and differentiation control in Saccharomyces cerevisiae (20). Type 2C protein phosphatases (PP2Cs) remove phosphate from Ser and Thr residues (21), and are widely represented in bacteria, fungi, plants, insects, and mammals (22, 23). In S. cerevisiae, there are seven PP2C-encoding genes (24–26), which share a conserved PP2C domain. Among them, Ptc2 serves to limit the maximum of activation of Hog1 (27). It also regulates negatively the unfolded-protein response through dephosphorylation of the Ser/Thr protein kinase Ire1 (28). In addition, Ptc2 and Ptc3 have been implicated in the regulation of progression through the cell cycle since they are capable of dephosphorylating Cdc28 at Thr169 (24), a residue essential for its activity as a cyclin-dependent kinase. Ptc2 in C. albicans shares a functional role with S. cerevisiae Ptc2, as a Captc2 mutant displays hypersensitivity to the genotoxic stress-inducing agents methyl methanesulfonate and hydroxyurea (29). It has been reported that Ppg1, a putative type 2A-related protein phosphatase (PP2A), is important for C. albicans filament extension, invasion, and virulence (30). Relatively little is known regarding PP2C in the regulation of hyphal development in response to the changing environments in C. albicans.
CO2 serves basic metabolic functions as both a building block and a waste product and thus plays a central role in the carbon cycle (31, 32). It is not a major component of the atmosphere, comprising only 0.0365%, but its levels are substantially higher in our bloodstream and tissues, where as an end product of respiration it is found at levels of roughly 5%. Elevated CO2 levels induce virulence factors such as capsule biosynthesis and filamentation in opportunistic fungal pathogens Cryptococcus neoformans (33, 34) and Candida albicans (35) through adenylyl cyclase-dependent signaling pathways (36, 37). CO2/HCO3− also signals independently of adenylyl cyclase to regulate levels of carbonic anhydrase (38, 39) and promote cell-fate transition (40). Here, we report that a high concentration of CO2 triggers the dephosphorylation of Ssn3 by Ptc2 that in turn reduces Ume6 phosphorylation and prevents its degradation, leading to sustained hyphal development. Our results demonstrate that the Ptc2-Ssn3 axis represents a new regulatory module of CO2 signaling.
RESULTS
Two distinct E3 ubiquitin ligases control Ume6 stability in response to hypoxia and high CO2, respectively.
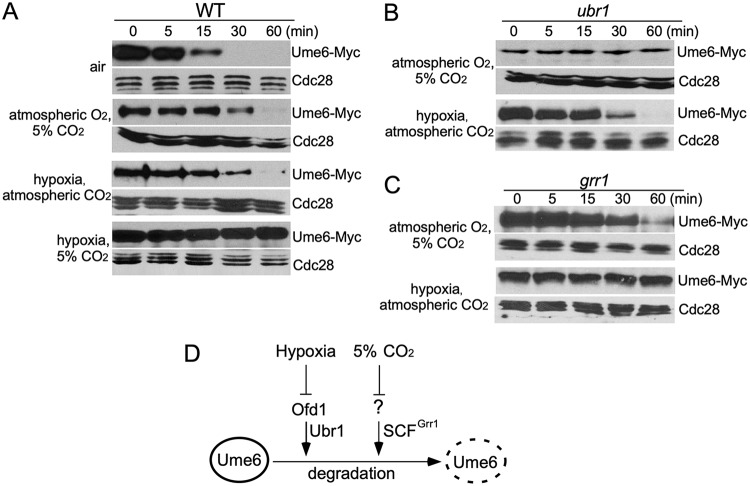
We have demonstrated that Ume6 was continuously degraded in air, but stable in hypoxia combined with 5% CO2 to promote hyphal elongation in C. albicans (17). Both low oxygen and high CO2 contributed to Ume6 stabilization, but neither alone was sufficient (Fig. 1A). Ofd1 regulates Ume6 stability in response to oxygen concentration (17). In Schizosaccharomyces pombe, Ofd1-mediated protein degradation in O2 requires the E3 ubiquitin ligase Ubr1 (41). To investigate whether a similar regulation exists in C. albicans, we examined Ume6 turnover in hypoxia (0.2% O2) or 5% CO2 by promoter shutoff assays. A gene encoding Ume6C778/785S, which had the Cys778 → Ser and Cys785 → Ser substitutions in the Gal4 DNA binding domain of Ume6, was expressed under the control of the MET3 promoter. The DNA binding domain of Ume6 was mutated in the construct to disrupt its affinity for DNA as MET3 expression could not be shut off completely when wild-type Ume6 was expressed (17). If Ubr1 is responsible for Ume6 degradation in O2, Ume6 is expected to be stable in atmospheric O2 plus 5% CO2. Indeed, we found that Ume6 was stable in the ubr1 mutant under this condition (Fig. 1B). Therefore, Ume6 degradation in atmospheric O2 depends on the Ubr1 ubiquitin ligase. Importantly, deletion of UBR1 could not block Ume6 degradation in atmospheric CO2 (Fig. 1B), suggesting that Ume6 degradation in response to CO2 concentration is controlled by additional E3 ligases. We have previously shown that stabilization of the hypha-specific G1-like cyclin Hgc1, like Ume6, requires both hypoxia and 5% CO2 (17). Hgc1 is unstable when expressed in S. cerevisiae, and Hgc1 degradation was blocked by deleting GRR1 (42), which encodes the F-box protein of the SCFGrr1 ubiquitin ligase complex. However, Hgc1 degradation was not blocked in C. albicans grr1 mutants in air (42, 43). We found that the C. albicans grr1 mutant hampered Hgc1 degradation only in hypoxia, and Hgc1 stability in the grr1 mutant was not affected by CO2 levels anymore (see Fig. S1 in the supplemental material). Thus, CO2-responsive Hgc1 degradation depends on the SCFGrr1. Given that both hyphal regulators, Ume6 and Hgc1, are stable in hypoxia plus 5% CO2, one would predict that the SCFGrr1 is also responsible for Ume6 degradation in response to CO2 level. As shown in Fig. 1C, the grr1 mutant blocked Ume6 degradation in hypoxia regardless of CO2 levels. The O2-sensitive and CO2-insensitive Ume6 stability in the grr1 mutant further supports the specificity of the SCFGrr1 for CO2-responsive Ume6 degradation. Together, our data suggest that Ume6 stability in C. albicans is regulated by two parallel E3 ubiquitin ligases under the control of specific signaling pathways in response to O2 and CO2, respectively (Fig. 1D).
FIG 1.
Two distinct E3 ubiquitin ligases control Ume6 degradation in response to O2 and CO2. (A) Ume6 stability was monitored by MET3 promoter shutdown assay. Wild-type C. albicans cells containing Ume6C778/785S-Myc (shown as Ume6-Myc) under the regulation of the MET3 promoter were grown in SCD (−Met, −Cys) for 2 h to induce their expression at room temperature. Twenty-five milliliters of medium was transferred from the culture to a petri dish (150 × 15 mm) and incubated in air, hypoxia (0.2% O2), 5% CO2, or hypoxia plus 5% CO2 as indicated at 30°C for 4 h. Methionine at 5 mM was then added to shut off the promoter. Aliquots were harvested at times indicated for the anti-Myc Western blot analysis. (B) Ubr1 is critical for O2-responsive Ume6 degradation. Ume6 stability was monitored by MET3 promoter shutdown assay in the ubr1 mutant under indicated conditions. (C) Ume6 degradation under atmospheric CO2 requires the SCFGrr1 E3 ligase. Ume6 stability was monitored by MET3 promoter shutdown assay in the grr1 mutant under indicated conditions. (D) Model for regulation of Ume6 degradation by oxygen and CO2. Ofd1 mediates the regulation of Ume6 degradation by E3 Ubr1 in response to oxygen, and CO2 regulates Ume6 degradation by SCFGrr1 through an unknown mechanism.
Hgc1 is stabilized in hypoxia in the grr1 mutant. Stability of Hgc1 was monitored by a MAL2 promoter shut down in air or in hypoxia (0.2% O2). Western blot analysis in C. albicans wild-type cells expressing Myc-Hgc1 from the MAL2 promoter. Download FIG S1, PDF file, 0.1 MB (131.4KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ptc2 is required for CO2-induced Ume6 stabilization.
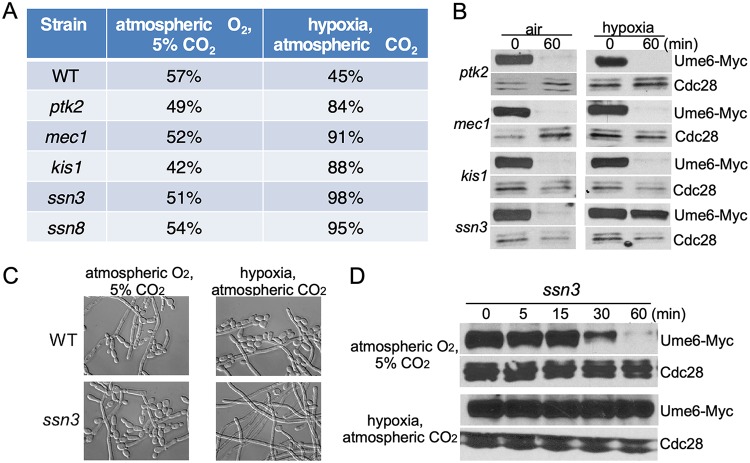
To identify the CO2 signaling pathway that regulates Ume6 stability, genetic screens were carried out to identify genes important for Ume6 stability in high CO2. We performed a screen with a knockout library of 674 unique genes in C. albicans (44) for mutants that are unable to sustain hyphal elongation in YEP+sucrose under hypoxia plus 5% CO2 (see Materials and Methods). The ptc2 mutant (type 2C protein phosphatase) was found to have impaired hyphal elongation under 5% CO2, but normal hyphal elongation in serum (Fig. 2A). Most of the ptc2 mutant cells converted to yeast under 5% CO2. However, about 50% cells of ptc2 mutant could form hyphae in hypoxia plus 5% CO2 (Fig. 2A), suggesting that deletion of PTC2 had no detectable defect in hypoxia-induced hyphal elongation. We also screened the GRACE library, a nonredundant library containing a total of 2,357 different mutants (45). ptc2 was the only mutant found specifically defective in CO2-induced hyphal elongation. These results indicated that Ptc2 is specifically required for hyphal elongation in high CO2. Correspondingly, Ume6 in the ptc2 mutant was unstable under hypoxia plus 5% CO2, less stable than in the WT strain under 5% CO2, and the same as in WT under hypoxia (Fig. 2B). The stability of Ume6 in the ptc2 mutant in 5% CO2 was similar to that in wild-type cells in air (Fig. 2B). Therefore, the defect of ptc2 mutant in Ume6 stability and hyphal elongation is detectable only in the presence of 5% CO2.
FIG 2.
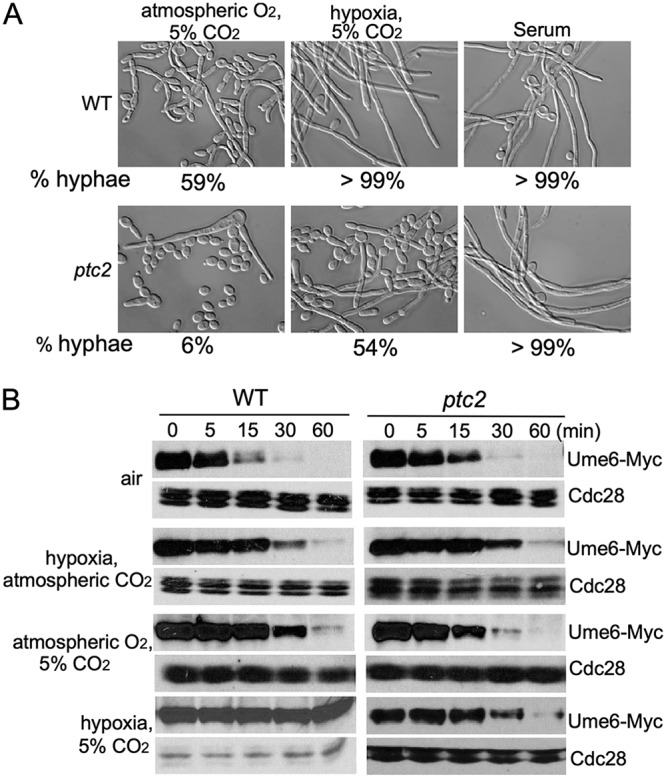
Ptc2 is critical for CO2-induced hyphal elongation. (A) Overnight cultures of wild-type and ptc2 mutant were diluted into YEPSucrose medium at 37°C. One-third of the samples were put into the CO2 incubator immediately, and cell morphology was examined after incubation for 5 h (left panels). One-third of the samples were put into the hypoxic chamber immediately and incubated for 12 h to analyze cell morphology in hypoxia plus 5% CO2 (middle panels). Ten percent serum was added to the other samples. Photographs were taken after 3.5 h of incubation (right panels). The percentage of cells forming hyphae was determined by counting at least 200 cells/sample. The data show the average from three independent experiments. The cells which had a length/width ratio of >4.5 and characteristic shape were considered hyphae. (B) Ume6 protein cannot be stabilized in 5% CO2 in ptc2 mutant. The protein stability of Ume6 in wild type and ptc2 mutant was monitored by MET3 promoter shutdown.
Ssn3 promotes Ume6 degradation in atmospheric CO2.
Ptc2 is a type 2C protein phosphatase (PP2C) in C. albicans (29). In S. cerevisiae, Ptc2 dephosphorylates Hog1, Ire1, and Cdc28, which are involved in the regulation of osmostress response, unfolded protein response, and cell cycle progression, respectively (24, 27, 28). So far, all known substrates of Ptc2 are kinases. Therefore, we hypothesize that Ptc2 inactivates a kinase to stabilize Ume6 in response to 5% CO2. To identify the kinase that promotes Ume6 degradation, we screened a knockout library of 80 kinases and kinase-related genes (46) for mutants that grew hyperfilamentously under 0.2% O2 but showed wild-type levels of filamentation under 5% CO2. Five mutants met the criteria in hyphal elongation (Fig. 3A). We then determined Ume6 stability in 4 putative kinase mutants under hypoxia, and the ssn3 mutant showed stabilization of Ume6 (Fig. 3B). Ssn3 is a cyclin-dependent protein kinase, and Ssn8 is the cyclin-like component for Ssn3 (19). They are components of the mediator complex (47). In hypoxia, hyphal elongation was fully maintained (Fig. 3C), and Ume6 was stable in the ssn3 mutant (Fig. 3D). Five percent CO2 had no effect on Ume6 stability and hyphal elongation in the ssn3 mutant (Fig. 3C and D, air versus 5% CO2). Our data indicated that Ssn3 is the kinase for Ume6 degradation in atmospheric CO2.
FIG 3.
Ssn3 is required for Ume6 degradation in atmospheric CO2. (A) The percentage of cells forming hyphae of indicated strains was determined as described for Fig. 2A. (B) The protein stability of Ume6 in indicated strains was monitored by MET3 promoter shutdown. (C) Morphology analysis of wild-type and ssn3 mutant cells was performed as described in Fig. 2A. (D) The protein stability of Ume6 in ssn3 mutant was monitored by MET3 promoter shutdown under 5% CO2 or hypoxia condition.
Phosphorylation at S437 by Ssn3 is required for CO2-regulated Ume6 degradation.
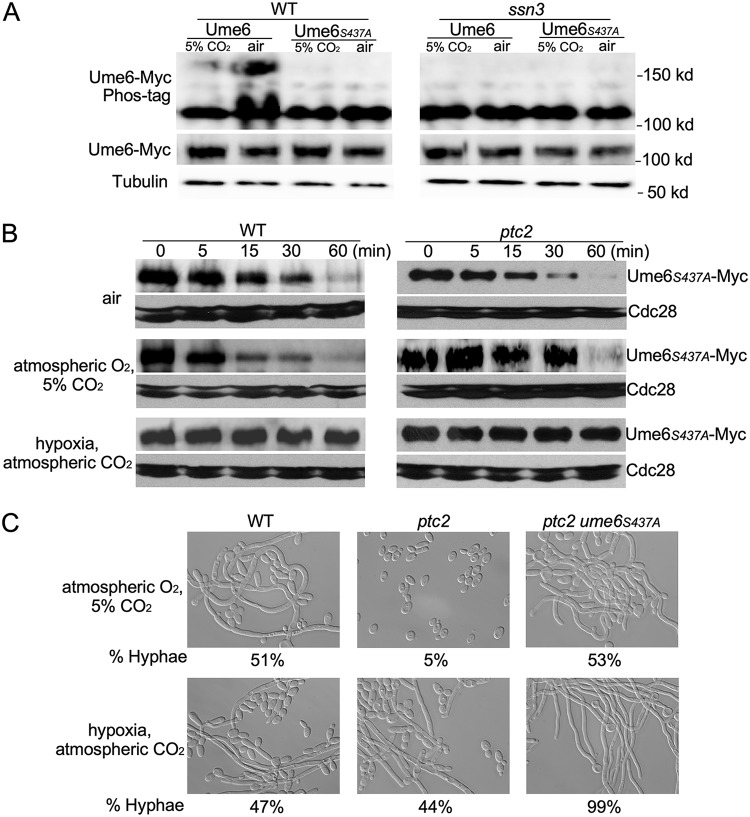
Given that the Ume6 degradation in response to CO2 requires the F-box protein Grr1, which is known to interact with phosphorylated targets (48), we predict that Ssn3 phosphorylates Ume6 in low CO2 to promote its degradation. Ume6 was expressed from its own promoter, and a significant portion of Ume6 showed a mobility shift in Phos-tag gels in WT cells in air, while very little Ume6 showed an upshift in 5% CO2 (Fig. 4A). Deletion of SSN3 abolished the mobility shift (Fig. 4A), indicating that the phosphorylation of Ume6 in response to atmospheric levels of CO2 is dependent on Ssn3. Using the GPS 2.0 (Group-Based Prediction System) phosphorylation prediction system (49) with Cdk8 as the kinase and threshold set to high, 8 S/T residues in Ume6 are predicted to be phosphorylation sites of Ssn3. Among them, S437 has the highest score, 10, and S440 has the second highest score, 9.66. We mutated S437 to Ala, and examined the Ume6S437A protein in Phos-tag gels. As shown in Fig. 4A, mobility shift was not observed with the Ume6S437A under both air and 5% CO2, suggesting that the S437 residue is a critical phosphorylation site by Ssn3 under atmospheric CO2. We next expressed Ume6S437A under the MET3 promoter to determine whether phosphorylation of S437 is essential for Ume6 degradation under atmospheric CO2. Using the promoter shutdown assay, Ume6S437A was partially stable in air (Fig. 4B) in comparison to Ume6 in air (Fig. 1A). Importantly, 5% CO2 did not increase the stability of Ume6S437A, while hypoxia was able to increase the stability of Ume6S437A (Fig. 4B). Therefore, the S437 is required for Ume6 phosphorylation and degradation under atmospheric CO2. We next examined Ume6S437A stability in the ptc2 mutant, in which Ume6 is degraded similarly in atmospheric CO2 and 5% CO2 (Fig. 2B). In contrast to wild-type Ume6 in the ptc2 mutant (Fig. 2B), Ume6S437A is similarly stable in atmospheric or 5% CO2 in the ptc2 mutant, and is completely stable in hypoxia (Fig. 4B). Thus, the Ser437-to-Ala mutation made Ume6 stable in atmospheric CO2, and bypassed the need for Ptc2. Next, we investigated whether the S437A mutation in Ume6 could rescue the defect of the ptc2 mutant in hyphal elongation under 5% CO2. We replaced both copies of UME6 with the Ume6S437A through Crispr-Cas9 (50). As shown in Fig. 4C, hyphal development could be sustained in the ptc2 ume6S437A mutant under hypoxia conditions. Therefore, the ume6S437A is epistatic to ptc2. We also mutated the S440 of Ume6 to Ala in the WT and ptc2 mutant by CRISPR-Cas9, but the UME6S440A did not affect hyphal elongation of the WT or ptc2 mutant under both atmospheric CO2 and 5% CO2 (Y. Lu, unpublished data). Therefore, phosphorylation of Ume6 at S437 by Ssn3 is critical for Ume6 degradation in atmospheric CO2. Our data suggest that Ptc2 and Ssn3 play opposite roles on the phosphorylation of Ume6 to control Ume6 stability and hyphal elongation in response to CO2.
FIG 4.
Mutating the Ssn3 phosphorylation site in Ume6 stabilizes Ume6 protein and sustains hyphal elongation under hypoxic condition. (A) Ume6 is phosphorylated at S437 in air in an Ssn3-dependent manner. Cells of wild type and ssn3 mutant carrying Ume6-Myc or Ume6S437A-Myc were collected in air or in 5% CO2 at 4 h. Protein was extracted for Phos-tag gel analysis. (B) Ume6S437A-Myc stability in wild-type and ptc2 mutant cells was monitored by MET3 promoter shutdown under indicated conditions. (C) Morphology analysis of indicated strains was performed as described in Fig. 2A.
Ssn3 is dephosphorylated in 5% CO2 in a Ptc2-dependent manner.
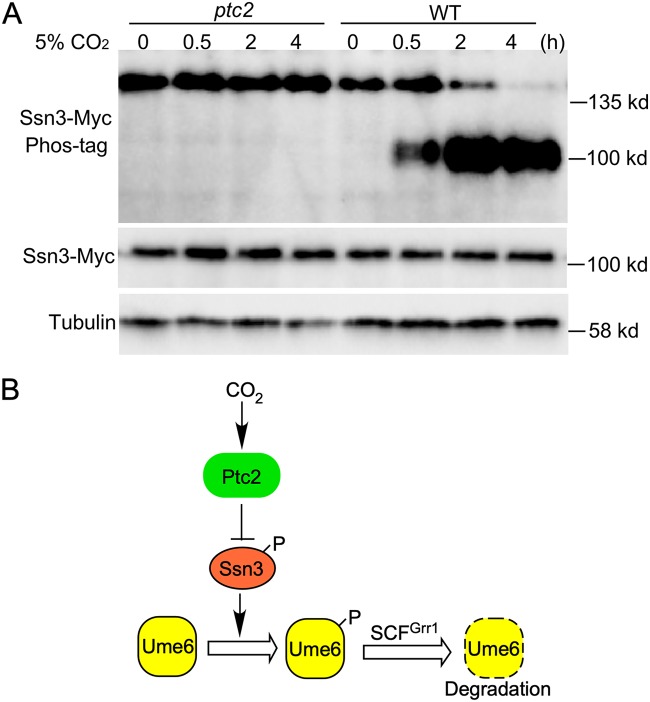
Since Ptc2 and Ssn3 play opposite roles on the regulation of hyphal elongation in response to CO2, we investigated how the CO2 signal is transduced to regulate Ume6 stability. In S. cerevisiae, Ptc2 is involved in the regulation of cell cycle progression as it is the main protein phosphatase acting to oppose the Cdk-activating kinase (CAK) on the activating phosphorylation site of CDK (Thr-169 of Cdc28) (24). Therefore, we hypothesize that Ptc2 inactivates Ssn3 by dephosphorylation in response to elevated CO2 to regulate Ume6 stability. To test this possibility, Ssn3-Myc was expressed from its own promoter in WT and ptc2 mutant cells. Five percent CO2 induced Ssn3 dephosphorylation in wild-type cells, as shown by the mobility shift in Phos-tag gels (Fig. 5A). In contrast, mobility shift was not observed in the ptc2 mutant in 5% CO2, indicating that Ptc2 is required for the dephosphorylation of Ssn3 in response to elevated CO2. Deletion of PTC2 had no effect on the phosphorylation of Ssn3 in air, as there is no band shift exhibited at the zero point between wild-type cells and the ptc2 mutant (Fig. 5A). Our data suggest that physiological levels of CO2 can induce Ptc2-mediated dephosphorylation of Ssn3. Hypophosphorylated Ssn3 fails to phosphorylate S437 in Ume6, resulting in stabilization of Ume6 to sustain hyphal development.
FIG 5.
Ssn3 is dephosphorylated in 5% CO2 in a Ptc2-dependent manner. (A) Cells of wild type and ptc2 mutant carrying Ssn3-Myc were collected at indicated time points after exposure to 5% CO2. Protein was extracted for Phos-tag gel analysis. (B) A schematic diagram depicting the CO2 signaling pathway mediated by the Ptc2-Ssn3 axis that controls hyphal elongation in C. albicans. A high level of CO2 triggers Ptc2 to dephosphorylate Ssn3. Phospho-Ssn3 promotes Ume6 phosphorylation, which leads to Ume6 degradation. The dashed circle represents degraded protein.
DISCUSSION
Sensing of CO2 and rapid adaptation to changing levels of CO2 are an essential process in all living cells. This is particularly important for pathogenic fungi that are able to grow in a wide range of CO2 levels from atmospheric 0.036% to physiological 5% in the hosts. CO2 is hydrolyzed into bicarbonate inside the cell naturally and through the activity of carbonic anhydrase when CO2 concentration is low. In C. albicans, adenylyl cyclase Cyr1 acts as a bicarbonate sensor to regulate hyphal morphogenesis. Through genetic screens, we identified a new CO2 signaling pathway that governs Ume6 protein stability to promote hyphal elongation in C. albicans. Ptc2, a type 2C protein phosphatase, is specifically required for CO2-responsive hyphal elongation. High levels of CO2 trigger Ptc2 to dephosphorylate Ssn3. The hypophosphorylated Ssn3 fails to phosphorylate Ume6 at the S437 residue, which prevents Ume6 from being targeted by SCFGrr1 for ubiquitination, and therefore stabilized to promote hyphal elongation (Fig. 5B). As we previously reported (17), the stabilization of Ume6 protein is coordinately regulated by hypoxia and high CO2. Here we demonstrate that Ume6 stability is controlled by two E3 ubiquitin ligases in response to hypoxia and high CO2, respectively. A recent study by Mendelsohn et al., showed that Cdk1-Hgc1 promotes Ume6 degradation via the SCFCdc4 ubiquitin ligase (51). Both SCFGrr1 and SCFCdc4 ubiquitin ligases likely participate in Ume6 degradation, as the Cacdc53ts mutant completely blocked Ume6 degradation while CDC4 shutdown only partially affected Ume6 degradation (51). Cdc53 is an essential protein of SCF complexes, including SCFGrr1 and SCFCdc4. Our promoter shutdown assay for Ume6 stability was carried out at 30°C with the Ume6-myc protein that is defective in DNA binding. Therefore, the expression level of hypha-induced genes, including HGC1, is expected to be low under our assay condition. Cdk1/Hgc1-activated Ume6 degradation may not contribute to Ume6 stability in this study. Like Ume6, Hgc1 degradation could also be regulated by multiple signaling pathways and E3 ubiquitin ligases (Fig. S1), which explains why the Cagrr1 deletion does not block Hgc1 degradation (42, 43). It is known that CO2 production is directly coupled to oxygen consumption of eukaryotic cells, and sites of hypoxia in vivo often contain increased levels of CO2. Therefore, our study may provide the underlying mechanism of how the interconnections and relationship are established between oxygen and carbon dioxide sensing with regard to fungal pathogenesis.
Our identification of the Ptc2-Ssn3 axis that governs CO2-responsive Ume6 stabilization and hyphal elongation provides molecular insights into fungal CO2 sensing. CO2 is a key determinant involved in fundamental biological processes, including growth, morphology, and virulence in fungi (52). Adenylyl cyclase acts as a bicarbonate sensor to promote hyphal growth in response to elevated CO2 levels in C. albicans (35). A recent report revealed the regulatory role of the TCA cycle in CO2 sensing and hyphal development through integration with the Ras1-cAMP signaling pathway in C. albicans (53). However, a pulse of activation of cAMP-PKA pathway promotes hyphal initiation, yet is not sufficient for long-lasting hyphal maintenance (14, 15). The Sch9 kinase has been shown to downregulate hyphal formation in hypoxia and high CO2 (54). Sch9 is also involved in the regulation of CO2-responsive carbonic anhydrase expression (39). However, hyperfilamentation of the sch9 mutant was detected only during growth on agar and at low temperature. Deletion of SCH9 resulted in even lower levels of hypha formation compared to wild-type cells in liquid media at 37°C (54), suggesting that Sch9 is unlikely to function through the same CO2 signaling pathway as Ssn3 in the regulation of Ume6 stability. Genome-wide analysis revealed that the transcription levels of a large number of genes are changed in C. albicans to adapt to high CO2. For example, genes related to the TCA cycle, genes responsive to stress and drugs, and amino acid synthesis-related genes are upregulated in 5% CO2. Whether these regulations occur through the Ptc2-Ssn3-mediated signaling pathway needs to be further investigated.
Ssn3, a cyclin-dependent kinase, promotes Ume6 degradation under atmospheric CO2 (Fig. 3). Under atmospheric CO2, Ume6 is phosphorylated in an Ssn3-dependent manner (Fig. 4A). Moreover, Ume6 is completely stable under atmospheric CO2 and hypoxia conditions in the ssn3 mutant (Fig. 3D) or when the Ssn3-dependent phosphorylation site (S437) is mutated (Fig. 4B). Based on these results, we propose that Ssn3 is inactivated in 5% CO2, thus preventing Ume6 from phosphorylating at S437, which is necessary for CO2-responsive Ume6 degradation. C. albicans mutants defective in the Ssn3 module of mediator lead to enhanced biofilm formation (55), and a nonsynonymous mutation in SSN3 is sufficient for regaining the ability to filament in the absence of Efg1 and Cph1 to damage macrophages (56). It is not clear if the regulatory roles of Ssn3 in filamentation in these two studies are through transcriptional regulation via the mediator complex or via regulation of Ume6 stability. In S. cerevisiae, a number of gene-specific transcriptional regulators have been defined as Ssn3 substrates, whose degradation is induced upon phosphorylation by Ssn3 (20). In response to nitrogen limitation, a decrease in Ssn3 levels leads to stabilization of two key transcription activators, Ste12 and Phd1, which promotes pseudohyphal growth in S. cerevisiae (57, 58). In this study, we did not observe a decrease in the protein level of Ssn3 when C. albicans cells were exposed to 5% CO2 (Fig. 5A), suggesting that inactivation of Ssn3 in 5% CO2 is not through downregulating SSN3 expression. A recent study reported that cyclin C (Ssn8) is destroyed in response to oxidative stress, leading to Cdk8 (Ssn3) inactivation (59). However, SSN8 expression is not regulated by CO2 levels, and overexpression of SSN8 had no effect on CO2-induced hyphal elongation (Y. Lu and H. Liu, unpublished data). Therefore, Ssn3 activity is probably not regulated through changing the levels of its associated cyclin Ssn8.
Our genetic screens identified a phosphatase Ptc2 and a kinase Ssn3 as the major positive and negative regulators in CO2 signaling of sustained hyphal development, respectively. Ptc2 is a type 2C Ser/Thr phosphatase that is conserved in eukaryotes and involved in a large variety of functional processes. Ptc2 dephosphorylates a number of kinases, including Hog1, Ire1, and Cdc28, to repress the activity of these kinases in S. cerevisiae (24, 27, 28). It is yet to be determined if Ptc2 regulates any of these kinases in C. albicans. Here we show that, in response to 5% CO2, Ssn3 is dephosphorylated in a Ptc2-dependent manner. Our data suggest that Ssn3 is a downstream target of Ptc2 in this CO2 signaling pathway, and Ssn3 activity is inhibited upon dephosphorylation by Ptc2. This study adds an additional layer of the regulation of Ssn3 activity and provides the first example, to our knowledge, of how Ssn3 activity is regulated through changing its phosphorylation state in response to environmental cues. Given that Ptc2-mediated inhibition of Ssn3 occurs through dephosphorylation, the origin of the activating phosphorylation should be considered. Two modes of Ssn3 activation can be envisaged: autophosphorylation or phosphorylation by upstream kinases. Ssn3 may be activated by autophosphorylation, as we did not identify another kinase mutant from our screening that exhibited a similar phenotype as the ssn3 mutant on the regulation of Ume6 stability. Such a PP2C-kinase regulatory module is also used by plants in ABA (abscisic acid) signaling, whereby ABA binding by PYR1/PYL/RCAR soluble ABA receptors inhibits PP2C phosphatases such as ABI1, ABI2, and HAB1 (60, 61), allowing serine-threonine SnRK2-type kinases (sucrose nonfermenting-1 [Snf1]-related protein kinase 2) to perform activation and phosphorylation of target proteins (62, 63). Taken together, our study elucidated a new regulatory mechanism for CO2 signaling in C. albicans through the Ptc2-Ssn3-medated protein phosphorylation/dephosphorylation system.
MATERIALS AND METHODS
Media and growth conditions.
C. albicans strains were routinely grown at 30°C in YPD (2% Bacto peptone, 2% dextrose, 1% yeast extract). Transformants were selected on synthetic medium (2% dextrose, 0.17% Difco yeast nitrogen base without ammonium sulfate, 0.5% ammonium sulfate, and auxotrophic supplements) or YPD + 200 µg/ml nourseothricin plates. Hyphal inductions were performed as follows. Strains were grown overnight in liquid YPD at 30°C, pelleted, washed twice in PBS, resuspended in an equal volume of PBS, and diluted 1:250 in YPSucrose medium (2% Bacto peptone, 2% sucrose, 1% yeast extract) with or without 10% serum at 37°C. For hyphal induction in hypoxia or 5% CO2, experiments were carried out using a Galaxy R170 CO2 incubator (Eppendorf). The oxygen and carbon dioxide concentrations were controlled by varying the concentration of nitrogen or carbon dioxide. Two hundred fifty microliters of prewarmed YPSucrose medium (buffered with citrate acid at pH 6.0) was added to each well of a 24-well plate, and 1 μl of overnight culture was inoculated into each well. The plate was placed into the incubator at 37°C immediately. After 12 h, cells were collected for morphological analysis.
Screening for mutant defective in hyphal elongation under 5% CO2.
The deletion mutant library affecting 674 genes of C. albicans (34) and the wild-type reference strain SN250 were grown overnight in liquid YPD at 30°C. Thirty-five mutants grew as elongated pseudohyphae, and they were excluded from further analysis. The remaining 639 mutants and wild-type cells were diluted at 1:250 to the buffered YPSucrose (pH 6.0) medium at 37°C under hypoxia (0.2% O2) plus 5% CO2 for 12 h. Thirty-three mutants were defective in hyphal elongation under this condition; only the ptc2 mutant was very defective in CO2-induced hyphal maintenance but had no defect in hyphal development in YPD + 10% serum.
Plasmid and strain construction.
The C. albicans strains used in this study are listed in Table S1 in the supplemental material. Primer sequences are listed in Table S2. The wild-type SN250 and ptc2 mutant were streaked on 5-fluoro-orotic acid-containing medium to generate Ura− strains. Two-step PCR was used to create pMET3-UME6C778/785S, S437A-13MYC. Two pairs of primers (primers 1 and 2 and primers 3 and 4) were used to PCR amplify overlapping UME6 fragments with the mutation in the overlapping region from the plasmid pMET3-UME6C778/785S (17). The resulting PCR products were purified and mixed as the templates for another round of PCR amplification using the primers 1 and 3, which produced the full-length UME6C778/785S, S437A sequence. The resulting mutant UME6C778/785S, S437A was inserted into the BamHI-MluI site of pPR673-MET3p (17) to generate pMET3-UME6C778/785S,S437A-13MYC by Gibson assembly. The plasmid was digested with PmlI within the MET3 promoter region for integration into the endogenous MET3 locus. Both copies of UME6 were replaced by UME6S437A using CRISPR-Cas9 (50) to construct C. albicans UME6S437A mutant strains as follows. The sgRNA (primers 5 and 6) was annealed to insert into pV1093 vector. The resulting plasmid was linearized by digestion with KpnI and SacI and was transformed into wild-type and ptc2 mutant cells with the repair template (primers 7 and 8). The mutants were verified by sequencing.
C. albicans strains used in this study. Download Table S1, PDF file, 0.3 MB (269.4KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.1 MB (106.6KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A 1.2-kb PCR product (primers 9 and 10) containing the C-terminal SSN3 coding region was inserted into the BamHI-MluI site of pPR673. The resulting plasmid was digested with SacI to target integration into its own locus to express Ssn3-13Myc.
Promoter shutdown assays.
C. albicans strains containing Ume6C778/785S-Myc (shown as Ume6-Myc) or Ume6C778/785S,S437A-Myc (shown as Ume6S437A-Myc) under the regulation of the MET3 promoter were grown in SCD (−Met, −Cys) for 2 h to induce their expression at room temperature. Twenty-five milliliters of medium was transferred from the culture to a petri dish (150 × 15 mm) and placed into air, a hypoxic chamber, or a CO2 incubator as indicated. After incubation at 30°C for 4 h, 5 mM methionine was added to shut off the promoter. Aliquots were collected after the times indicated, and protein levels were analyzed via Western blotting.
Phos-tag SDS-PAGE.
Phosphorylation states of Ume6-Myc and Ssn3-Myc were examined using Phos-tag SDS-PAGE, which is a phospho-affinity SDS-PAGE developed by Kinoshita et al. (64). Phos-tag acrylamide was purchased from Wako Chemicals (Osaka, Japan). Separating gels were made by copolymerization of acrylamide with Phos-tag acrylamide. Phos-tag SDS-PAGE was performed on 6% polyacrylamide gels containing 50 μM Phos-tag acrylamide and 100 μM MnCl2 in 10-mA current at 4°C. The separated proteins were transferred to PVDF membranes. Immunoreaction of the membrane was then carried out.
ACKNOWLEDGMENTS
We thank G. R. Fink for plasmids and S. Noble and A. Mitchell via the Fungal Genetics Stock Center for C. albicans deletion collections.
This work was supported by the National Natural Science Foundation of China grant 31770162 to Y.L. and grant 31700133 to C.S. and supported by the National Institutes of Health grants R01GM117111-A1 and R01AI099190 to H.L.
Y.L., C.S., and H.L. designed the research; Y.L., C.S., S.R., and Y.Y. performed the research; Y.L., C.S., and H.L. analyzed the data; Y.L., C.S., and H.L. wrote the paper.
Footnotes
Citation Lu Y, Su C, Ray S, Yuan Y, Liu H. 2019. CO2 signaling through the Ptc2-Ssn3 axis governs sustained hyphal development of Candida albicans by reducing Ume6 phosphorylation and degradation. mBio 10:e02320-18. https://doi.org/10.1128/mBio.02320-18.
Contributor Information
David Kadosh, University of Texas Health Science Center at San Antonio.
Michael Lorenz, University of Texas Health Science Center.
REFERENCES
- 1.Noble SM, Gianetti BA, Witchley JN. 2017. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen ID, Wilson D, Wachtler B, Brunke S, Naglik JR, Hube B. 2012. Candida albicans dimorphism as a therapeutic target. Expert Rev Anti Infect Ther 10:85–93. doi: 10.1586/eri.11.152. [DOI] [PubMed] [Google Scholar]
- 4.Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Curr Opin Microbiol 10:314–319. doi: 10.1016/j.mib.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romani L, Bistoni F, Puccetti P. 2003. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr Opin Microbiol 6:338–343. doi: 10.1016/S1369-5274(03)00081-X. [DOI] [PubMed] [Google Scholar]
- 6.Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 8.Schaller M, Korting HC, Schafer W, Bastert J, Chen W, Hube B. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol 34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 9.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Forster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Hader A, Kurzai O, Luo T, Kruger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19:1354–1365. doi: 10.1091/mbc.e07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res 9:126–142. doi: 10.1111/j.1567-1364.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, Wang Y, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A 106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Su C, Unoje O, Liu H. 2014. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci U S A 111:1975–1980. doi: 10.1073/pnas.1318690111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Su C, Liu H. 2012. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog 8:e1002663. doi: 10.1371/journal.ppat.1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Su C, Solis NV, Filler SG, Liu H. 2013. Synergistic regulation of hyphal elongation by hypoxia, CO(2), and nutrient conditions controls the virulence of Candida albicans. Cell Host Microbe 14:499–509. doi: 10.1016/j.chom.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes BT, Espenshade PJ. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J 27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchin S, Yeghiayan P, Carlson M. 1995. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci U S A 92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemet J, Jelicic B, Rubelj I, Sopta M. 2014. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 97:22–27. doi: 10.1016/j.biochi.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Cohen P. 1989. The structure and regulation of protein phosphatases. Annu Rev Biochem 58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 22.Schweighofer A, Hirt H, Meskiene I. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Lammers T, Lavi S. 2007. Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit Rev Biochem Mol Biol 42:437–461. doi: 10.1080/10409230701693342. [DOI] [PubMed] [Google Scholar]
- 24.Cheng A, Ross KE, Kaldis P, Solomon MJ. 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev 13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L, Whiteway M, Ramos C, Rodriguez-Medina JR, Shen SH. 2002. The YHR076w gene encodes a type 2C protein phosphatase and represents the seventh PP2C gene in budding yeast. FEBS Lett 527:323–325. doi: 10.1016/S0014-5793(02)03247-7. [DOI] [PubMed] [Google Scholar]
- 26.Ruan H, Yan Z, Sun H, Jiang L. 2007. The YCR079w gene confers a rapamycin-resistant function and encodes the sixth type 2C protein phosphatase in Saccharomyces cerevisiae. FEMS Yeast Res 7:209–215. doi: 10.1111/j.1567-1364.2006.00160.x. [DOI] [PubMed] [Google Scholar]
- 27.Young C, Mapes J, Hanneman J, Al-Zarban S, Ota I. 2002. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot Cell 1:1032–1040. doi: 10.1128/EC.1.6.1032-1040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. 1998. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol 18:1967–1977. doi: 10.1128/MCB.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, Zhao J, Li J, Zhang L, Jiang L. 2010. Functional characterization of the PP2C phosphatase CaPtc2p in the human fungal pathogen Candida albicans. Yeast 27:753–764. doi: 10.1002/yea.1778. [DOI] [PubMed] [Google Scholar]
- 30.Albataineh MT, Lazzell A, Lopez-Ribot JL, Kadosh D. 2014. Ppg1, a PP2A-type protein phosphatase, controls filament extension and virulence in Candida albicans. Eukaryot Cell 13:1538–1547. doi: 10.1128/EC.00199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hetherington AM, Raven JA. 2005. The biology of carbon dioxide. Curr Biol 15:R406–R410. doi: 10.1016/j.cub.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 32.Janssen PJ, Lambreva MD, Plumere N, Bartolucci C, Antonacci A, Buonasera K, Frese RN, Scognamiglio V, Rea G. 2014. Photosynthesis at the forefront of a sustainable life. Front Chem 2:36. doi: 10.3389/fchem.2014.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahn YS, Cox GM, Perfect JR, Heitman J. 2005. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol 15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 34.Alspaugh JA, Perfect JR, Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Muhlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. 2002. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilera J, Petit T, de Winde JH, Pronk JT. 2005. Physiological and genome-wide transcriptional responses of Saccharomyces cerevisiae to high carbon dioxide concentrations. FEMS Yeast Res 5:579–593. doi: 10.1016/j.femsyr.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Pohlers S, Martin R, Kruger T, Hellwig D, Hanel F, Kniemeyer O, Saluz HP, Van Dijck P, Ernst JF, Brakhage A, Muhlschlegel FA, Kurzai O. 2017. Lipid signaling via Pkh1/2 regulates fungal CO2 sensing through the kinase Sch9. mBio 8:e02211-16. doi: 10.1128/mBio.02211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du H, Guan G, Xie J, Cottier F, Sun Y, Jia W, Muhlschlegel FA, Huang G. 2012. The transcription factor Flo8 mediates CO2 sensing in the human fungal pathogen Candida albicans. Mol Biol Cell 23:2692–2701. doi: 10.1091/mbc.E12-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CY, Yeh TL, Hughes BT, Espenshade PJ. 2011. Regulation of the Sre1 hypoxic transcription factor by oxygen-dependent control of DNA binding. Mol Cell 44:225–234. doi: 10.1016/j.molcel.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang A, Lane S, Tian Z, Sharon A, Hazan I, Liu H. 2007. Temporal and spatial control of HGC1 expression results in Hgc1 localization to the apical cells of hyphae in Candida albicans. Eukaryot Cell 6:253–261. doi: 10.1128/EC.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li WJ, Wang YM, Zheng XD, Shi QM, Zhang TT, Bai C, Li D, Sang JL, Wang Y. 2006. The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol Microbiol 62:212–226. doi: 10.1111/j.1365-2958.2006.05361.x. [DOI] [PubMed] [Google Scholar]
- 44.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 46.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog 6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balciunas D, Ronne H. 1995. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res 23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol Cell Biol 21:2506–2520. doi: 10.1128/MCB.21.7.2506-2520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. 2008. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendelsohn S, Pinsky M, Weissman Z, Kornitzer D. 2017. Regulation of the Candida albicans hypha-inducing transcription factor Ume6 by the CDK1 cyclins Cln3 and Hgc1. mSphere 2:e00248-16. doi: 10.1128/mSphere.00248-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bahn YS, Muhlschlegel FA. 2006. CO2 sensing in fungi and beyond. Curr Opin Microbiol 9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Tao L, Zhang Y, Fan S, Nobile CJ, Guan G, Huang G. 2017. Integration of the tricarboxylic acid (TCA) cycle with cAMP signaling and Sfl2 pathways in the regulation of CO2 sensing and hyphal development in Candida albicans. PLoS Genet 13:e1006949. doi: 10.1371/journal.pgen.1006949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stichternoth C, Fraund A, Setiadi E, Giasson L, Vecchiarelli A, Ernst JF. 2011. Sch9 kinase integrates hypoxia and CO2 sensing to suppress hyphal morphogenesis in Candida albicans. Eukaryot Cell 10:502–511. doi: 10.1128/EC.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindsay AK, Morales DK, Liu Z, Grahl N, Zhang A, Willger SD, Myers LC, Hogan DA. 2014. Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS Genet 10:e1004567. doi: 10.1371/journal.pgen.1004567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wartenberg A, Linde J, Martin R, Schreiner M, Horn F, Jacobsen ID, Jenull S, Wolf T, Kuchler K, Guthke R, Kurzai O, Forche A, d’Enfert C, Brunke S, Hube B. 2014. Microevolution of Candida albicans in macrophages restores filamentation in a nonfilamentous mutant. PLoS Genet 10:e1004824. doi: 10.1371/journal.pgen.1004824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson C, Goto S, Lund K, Hung W, Sadowski I. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 58.Raithatha S, Su TC, Lourenco P, Goto S, Sadowski I. 2012. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol Cell Biol 32:664–674. doi: 10.1128/MCB.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stieg DC, Willis SD, Ganesan V, Ong KL, Scuorzo J, Song M, Grose J, Strich R, Cooper KF. 2018. A complex molecular switch directs stress-induced cyclin C nuclear release through SCF(Grr1)-mediated degradation of Med13. Mol Biol Cell 29:363–375. doi: 10.1091/mbc.E17-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Li J, Peterson FC, Jensen DR, Yong EL, Volkman BF, Cutler SR, Zhu JK, Xu HE. 2009. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 62.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hgc1 is stabilized in hypoxia in the grr1 mutant. Stability of Hgc1 was monitored by a MAL2 promoter shut down in air or in hypoxia (0.2% O2). Western blot analysis in C. albicans wild-type cells expressing Myc-Hgc1 from the MAL2 promoter. Download FIG S1, PDF file, 0.1 MB (131.4KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. albicans strains used in this study. Download Table S1, PDF file, 0.3 MB (269.4KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.1 MB (106.6KB, pdf) .
Copyright © 2019 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.