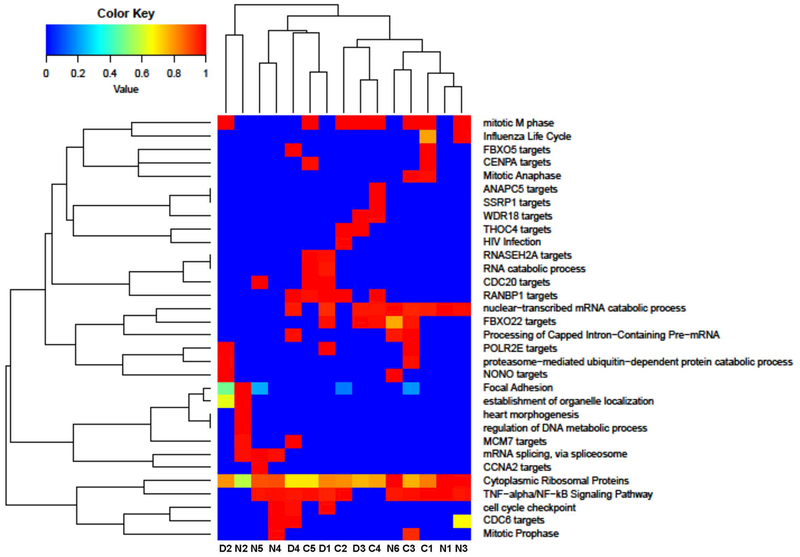
Figure 6: Comparisons of similarity of activated pathways among samples in normal controls (C1–5), CGD patients (D1–4) and of IFNGRD patients (N1–6).
It was possible to detect that Cytoplasmatic Ribossomal proteins, mRNA processing, Type II Interferon signaling (IFNG), EGFR1 Signaling pathway, TGF-beta Receptor Signaling pathway and Insulin Signaling are present in different intensities in all samples.