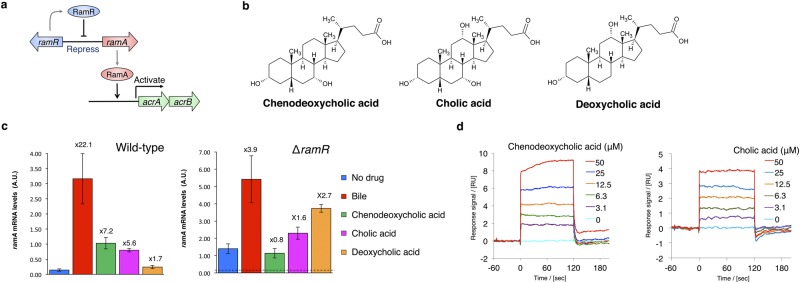
Figure 1.
Chenodeoxycholic and cholic acids activate the ramA gene in a RamR-dependent manner. (a) Model for gene regulation pathway by RamR. RamR represses the ramA gene encoding the activator protein for the acrAB drug efflux pump genes. RamR binds to the region between the ramR and ramA genes, while RamA binds to the upstream region to acrAB. (b) The chemical structures of chenodeoxycholic, cholic and deoxycholic acids. (c) Effect of bile and chenodeoxycholic, cholic and deoxycholic acids on the expression of ramA in wild-type and ∆ramR Salmonella strains, as assessed by qRT-PCR. Cells were grown in LB broth supplemented with 25.6 mg/ml bile, or 5 mM of chenodeoxycholic, cholic or deoxycholic acids. Values above the bars indicate the fold difference in ramA mRNA levels relative to the control in the same strain (wild-type or ∆ramR). In the right panel (expression in ∆ramR), the horizontal broken lines represents ramA mRNA levels in the wild-type control. The error bars indicate the standard deviation from three independent replicates. A.U., arbitrary units. (d) Binding of chenodeoxycholic and cholic acids to RamR detected by SPR analysis. RamR was immobilized onto a sensor chip and then the chenodeoxycholic or cholic acid was passed over the sensor surface at the various concentrations indicated. A representative result from one of three experiments that produced similar data is shown. SPR, surface plasmon resonance.