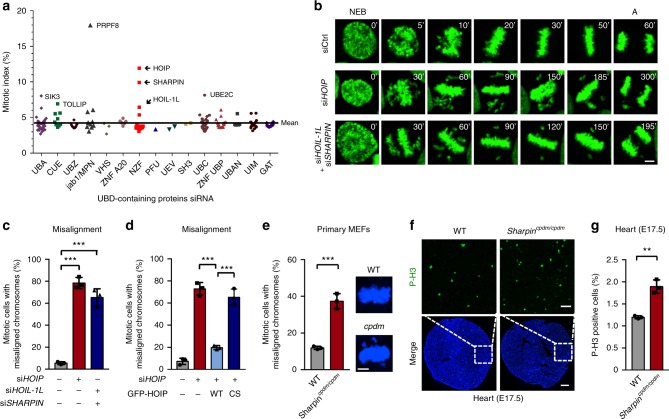
Fig. 1.
The linear ubiquitin ligase complex LUBAC is required for mitotic progression. a The mitotic index of HeLa cells transfected with an arrayed siRNA library targeting 190 genes encoding UBD-containing proteins. Full line, the mean value (4.2%). UBD, ubiquitin-binding domain. Proteins containing two or more UBDs were displayed once. b Time-lapse images of control or LUBAC depleted HeLa/GFP-H2B cells. The numbers are minutes after NEB. NEB nuclear envelope breakdown; A anaphase. Scale bar, 5 μm. c The percentage of mitotic cells with misaligned chromosomes in b. sicontrol, n = 349 cells; siHOIP, n = 340 cells; siHOIL-1L + siSHARPIN, n = 377 cells. d Expression of GFP-HOIP wild type (WT), but not its ligase-dead mutant (CS), rescued chromosome misalignment in HOIP depleted HeLa/RFP-H2B cells. sicontrol, n = 118 cells; siHOIP, n = 181 cells; HOIP WT + siHOIP, n = 175 cells; HOIP CS + siHOIP, n = 175 cells. e The percentage of mitotic cells with misaligned chromosomes in primary MEF cells from E13.5 embryos of wild-type or Sharpincpdm/cpdm mice. WT, n = 109 cells; cpdm, n = 130 cells. Scale bar, 5 μm. f Confocal microscope images of sections of E17.5 heart tissues of wild-type or Sharpincpdm/cpdm mice stained with antibody against Pho-H3. Top panels are enlargements of phospho-H3 staining of the regions outlined by white dashed boxes. Green, P-H3; blue, DNA. Scale bars, 200 μm (main image) and 50 μm (magnified region). g Quantification of P-H3 positive cells of the entire section in f. P-H3-positive cells were calculated automatically by the Volocity 6.0 software. n = 6 embryos per group. Data are presented as mean ± s.d. of three independent experiments (c, d, e and g). **P < 0.01, ***P < 0.001; two-sided Student’s t-test