Abstract
Chromobox 6 (CBX6) is a subunit of Polycomb Repressive Complex 1 (PRC1) that mediates epigenetic gene repression and acts as an oncogene or tumor suppressor in a cancer type-dependent manner. The specific function of CBX6 in breast cancer is currently undefined. In this study, a comprehensive analysis of The Cancer Genome Atlas (TCGA) dataset led to the identification of CBX6 as a consistently downregulated gene in breast cancer. We provided evidence showing enhancer of zeste homolog 2 (EZH2) negatively regulated CBX6 expression in a Polycomb Repressive Complex 2 (PRC2)-dependent manner. Exogenous overexpression of CBX6 inhibited cell proliferation and colony formation, and induced cell cycle arrest along with suppression of migration and invasion of breast cancer cells in vitro. Microarray analyses revealed that CBX6 governs a complex gene expression program. Moreover, CBX6 induced significant downregulation of bone marrow stromal cell antigen-2 (BST2), a potential therapeutic target, via interactions with its promoter region. Our collective findings support a tumor suppressor role of CBX6 in breast cancer.
Introduction
Polycomb group proteins (PcG) are important epigenetic regulators that function to maintain transcriptional repression. These proteins assemble into two major complexes in mammals, designated Polycomb Repressive Complex 1 (PRC1) and 2 (PRC2). The core components of the PRC2 complex include EZH1/2, SUZ12, EED, and RBAP46/48. Several other cofactors, such as JARID2, AEBP2 and Polycomb-like proteins (PCL1/PHF1, PCL2/MTF2, PCL3/PHF19) act as recruiters or modulators of PRC2 enzymatic activity1–4. PRC2 catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3), a marker of transcriptional repression, via its methyltransferase subunits EZH1/25. However, the components of PRC1 are considerably heterogeneous. For example, each of the Drosophila subunits has several homologs in mammals that assemble to form different PRC1 complex types. The core components of the canonical PRC1 complex include RING1A/B, PCGF, CBX, and PHC6. PRC1 monoubiquitinates histone H2A at lysine 119 (H2AK119ub) through the E3 ligase activity of RING1A/B, thereby contributing to gene silencing7. PRC2 and PRC1 are proposed to interact with each other to maintain gene repression. Canonically, PRC2 ‘writes’ H3K27me3 on chromatin of a given target gene locus, followed by binding of PRC1 to H3K27me3, leading to monoubiquitylation of H2A and subsequent chromatin compaction, and ultimately, gene repression8. Recent studies have shown that PRC1 can be recruited to target loci in a H3K27me3-independent manner and PRC1-dependent H2AK119ub1 recruits PRC2 to target genes6,9. PcG proteins are involved in multiple biological processes, including maintenance of cell identity, differentiation, proliferation, and cancer progression10–15.
Drosophila Polycomb protein (Pc) binds to H3K27me3 through a conserved N-terminal chromodomain16. Five orthologues of Drosophila Pc exist in mammals (CBX2, CBX4, CBX6, CBX7 and CBX8). Accumulating evidence supports critical roles of CBX proteins in tumorigenesis17–19. Remarkably, CBX proteins can act as either oncogenes or tumor suppressors in different cancer types. For example, CBX7 functions as a tumor suppressor and its expression is negatively associated with increased malignancy grades in bladder, pancreatic, glioma, breast, gastric, and colon carcinomas20. Conversely, CBX7 is overexpressed in prostate and ovarian cancer, implying an oncogenic role in these cancer types20. CBX8 acts as an oncogene in hepatocellular carcinoma (HCC) and promotes tumor growth and metastasis via activation of AKT/β-catenin signaling21, but suppresses cell migration, invasion and metastasis in esophageal squamous cell carcinoma (ESCC) and inhibits epithelial-mesenchymal transition (EMT) by repressing SNAIL expression22. The results of our primary study suggest that CBX6 is downregulated in glioblastomas and its overexpression reduces cell proliferative capacity23. However, frequent upregulation of CBX6 in HCC in association with promotion of cancer cell growth, both in vitro and in vivo, and poor prognosis has also been reported24. Therefore, the functions of individual CBX proteins in any cancer type should be investigated separately. While a number of CBX proteins, such as CBX2, 4, 7, and 8, have been shown to play vital roles in breast cancer progression25–28, the specific function of CBX6 in breast cancer progression remains to be elucidated.
Experiments from the current study demonstrated that CBX6 expression was frequently downregulated in breast cancer. Notably, CBX6 was silenced epigenetically by EZH2 in a PRC2-dependent manner. In functional analyses, overexpression of CBX6 resulted in cell proliferation inhibition, induced cell cycle arrest and dramatically suppressed the migration and invasion capacities of MCF-7 cells. Furthermore, CBX6 induced significant downregulation of BST2 via binding to its promoter region to exert potential antitumor activity.
Results
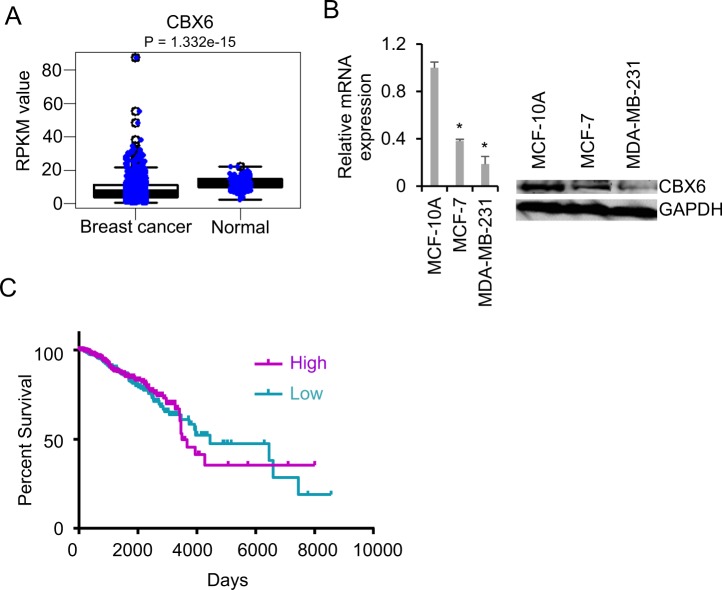
CBX6 is frequently downregulated in human breast cancer
To determine the specific role of CBX6 in breast cancer, we comprehensively analyzed The Cancer Genome Atlas (TCGA) dataset for aberrant expression of this gene (GSE62944). Significant downregulation of CBX6 was observed in breast cancer tissues compared with controls, as shown in Fig. 1A. Gene expression profiling experiments have facilitated the identification of several subtypes of breast cancer, including luminal A, luminal B, HER2-enriched, and basal-like. Examination of the TCGA dataset revealed that CBX6 is not differentially expressed in different subtypes of breast cancer (Supplementary Fig. S1A). CBX6 expression was further analyzed in breast cancer samples with different histological grades. Our data showed similar expression profiles of CBX6 at different stages (Supplementary Fig. S1B). To extend these observations, we tried to examine the expression of CBX6 by immunohistochemistry (IHC) in normal breast and breast cancer tissues. The signals detected using the CBX6 antibody (Millipore 09-030) are mainly located in the cytoplasm and connective tissues (Supplementary Fig. S2A). We interpreted that the IHC signal generated from this antibody was nonspecific, because CBX6 is primarily a nuclear protein as revealed by the immunofluorescence analysis of GFP-CBX6 fusion in MCF-7 cells (Supplementary Fig. S2B). The antibody recognized CBX6 immunoprecipitated from cell lysates (Supplementary Fig. S2C), and a band at the correct molecular weight of CBX6 in total cell lysates, but showed cross-reactivity with nonspecific bands of higher molecular weight. Next, the expression of CBX6 was assessed by qRT-PCR and by Western blotting using the antibody (Millipore 09-030) in a human non-tumorigenic epithelial cell line, MCF-10A, and two human breast adenocarcinoma cell lines, MCF-7 and MDA-MB-231. Consistently, CBX6 was significantly downregulated in breast cancer cells, compared with non-tumorigenic epithelial cells (Fig. 1B). In view of these findings, the association between CBX6 levels and clinical progression of breast cancer was further explored. Kaplan-Meier survival analysis of the TCGA breast cancer dataset showed the overall survival time did not significantly differ between patients with high CBX6 expression and low CBX6 expression when bifurcating gene expression at the median (HR = 0.94, P = 0.61) (Fig. 1C). However, survival analysis by the Kaplan-Meier plotter29, which curated the survival and gene expression data (Affymetrix microarray platform) of 5,143 breast cancer patients deposited in GEO29, showed patients with high CBX6 expression displayed significantly longer overall survival (OS) (HR = 0.78, P = 0.021) and recurrence-free survival (RFS) (HR = 0.73, P = 2.7e-08) than those with lower CBX6 expression30 (Supplementary Fig. S3). The discrepancy between the two datasets suggests the correlation between CBX6 expression and breast cancer patient survival needs further study.
Figure 1.
Chromobox 6 (CBX6) is downregulated in breast cancer. (A) mRNA expression of CBX6 in breast cancer versus normal breast tissues. The data are retrieved from the RNA-Seq datasets of The Cancer Genome Atlas (TCGA). The expression values are presented as Reads Per Kilobase of transcript per Million mapped reads (RPKM). An unpaired two-tailed t-test was used to evaluate significant differences. P values are indicated at the top of the graph. (B) CBX6 expression in human non-tumorigenic breast epithelial (MCF-10A) and two human breast adenocarcinoma (MCF-7 and MDA-MB-231) cell lines. Left: qRT-PCR analysis of CBX6 mRNA level in MCF-10A, MCF-7 and MDA-MB-231 cells. GAPDH was used for normalization of expression. Data are presented as mean ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right: Western blot analysis of CBX6 protein levels in MCF-10A, MCF-7 and MDA-MB-231 cells with GAPDH as a loading control. (C) Kaplan-Meier analysis of overall survival of breast cancer patients with different levels of CBX6 expression. Patient survival and gene expression data were downloaded from TCGA’s data portal and patients were split into two groups with the median CBX6 mRNA expression level as the cut-off.
EZH2 negatively regulates CBX6 in breast cancer
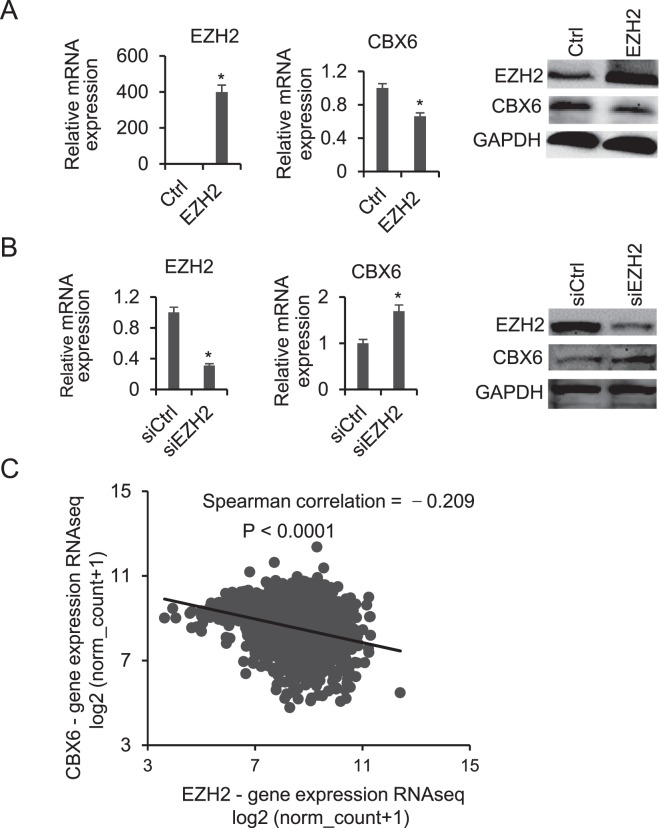
The mechanisms underlying CBX6 downregulation in breast cancer are not known at present. To identify the specific regulatory factors of CBX6 expression, we data-mined the ChIP-sequencing (ChIP-Seq) datasets deposited in the Gene Expression Omnibus (GEO). Interestingly, EZH2 and H3K27me3 peaks were detected at the CBX6 promoter in human mammary epithelial cells (HMEC) (Supplementary Fig. S4A), and a negative correlation between EZH2 and CBX6 expression was observed in MCF-10A, MCF-7, and MDA-MB-231 cells (Supplementary Fig. S4B). EZH2, which is highly expressed in breast cancer, promotes tumor progression and is associated with poorer patient outcome31. To further ascertain whether EZH2 is involved in regulation of CBX6, we performed overexpression and knockdown experiments in MCF-7 cells. CBX6 levels were markedly decreased upon EZH2 overexpression (Fig. 2A). Conversely, knockdown of EZH2 led to significant upregulation of CBX6 (Fig. 2B). Examination of the TCGA dataset further confirmed the negative correlation between EZH2 and CBX6 expression patterns in breast cancer (Spearman r = −0.209, P < 0.0001) (Fig. 2C).
Figure 2.
CBX6 is negatively regulated by EZH2. (A) Overexpression of EZH2 leads to downregulation of CBX6. Left and middle panels: qRT-PCR analysis of EZH2 and CBX6 mRNA in control (empty vector) and EZH2-overexpressing cells using GAPDH for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right panel: Western blot analysis of EZH2 and CBX6 protein levels in control (empty vector) and EZH2-overexpressing cells using GAPDH as a loading control. MCF-7 cells were transiently transfected with an empty or EZH2 vector for 48 hours, then cells were harvested, and total RNA and proteins were extracted for analysis. (B) Knockdown of EZH2 using siRNA leads to upregulation of CBX6. Left and middle panels: qRT-PCR analysis of EZH2 and CBX6 mRNA levels in siRNA control and EZH2 knockdown cells using GAPDH for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right: Western blot analysis of protein levels in siRNA control and EZH2 knockdown cells using GAPDH as a loading control. MCF-7 cells were transfected with control siRNA or siEZH2, total RNA and protein were extracted 48 h post-transfection for analysis. (C) Spearman’s rank correlation coefficient indicating a negative correlation between CBX6 and EZH2 mRNA levels in breast cancer tissues based on The Cancer Genome Atlas (TCGA) RNA-Seq data.
Regulation of CBX6 expression by EZH2 is PRC2-dependent
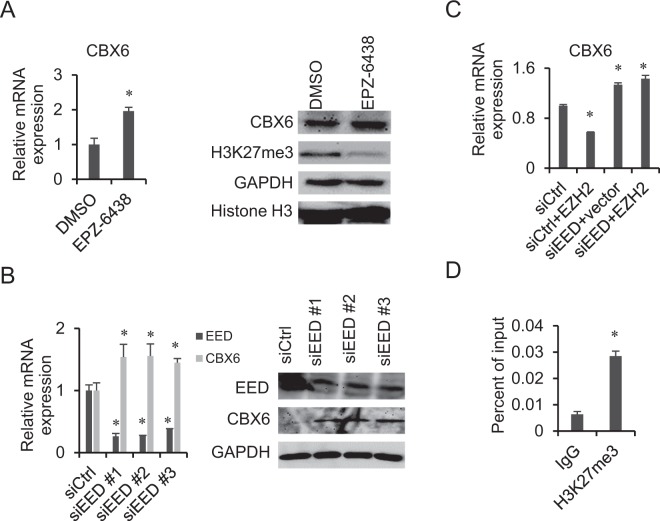
The gene regulatory activity of EZH2 can either be PCR2-dependent or -independent. Accordingly, we investigated whether or not EZH2-mediated downregulation of CBX6 is influenced by PRC2. To this end, MCF-7 cells were treated with the EZH2 inhibitor, EPZ-6438. As shown in Fig. 3A, following EPZ-6438 treatment, the H3K27me3 level was dramatically decreased and that of CBX6 was significantly increased in MCF-7 cells. EED, a core component of PRC2, is indispensable for PRC2 activity. We employed three different siRNAs for EED knockdown in MCF-7 cells, which all led to a marked increase in CBX6 expression (Fig. 3B). In addition, EZH2 overexpression no longer affected CBX6 expression after EED knockdown in MCF-7 cells (Fig. 3C). Furthermore, H3K27me3 enrichment at the CBX6 promoter was detected using ChIP-qPCR (Fig. 3D), indicating regulation of CBX6 expression by EZH2 is PRC2-dependent.
Figure 3.
EZH2 negatively regulates CBX6 expression in a PRC2-dependent manner. (A) Treatment with the EZH2 inhibitor, EPZ-6438, induces downregulation of CBX6 in MCF-7 cells. Left: qRT-PCR analysis of CBX6 mRNA in MCF-7 cells treated with DMSO or EPZ-6438. GAPDH was used for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right: Western blot analysis. MCF-7 cells were treated with 50 µM EPZ-6438 or DMSO for 5 days, then total RNA and protein were extracted for analysis. (B) Knockdown of EED is associated with upregulation of CBX6 in MCF-7 cells. Left: qRT-PCR analysis of CBX6 mRNA in siRNA control (siCtrl) and EED knockdown cells using GAPDH for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right: Western blot analysis of siRNA control and EED knockdown cells with indicated antibodies. GAPDH was used as the loading control. MCF-7 cells were transfected with siEED or control siRNA at a final concentration of 50 nM using the Lipofectamine RNAiMAX transfection reagent. Total RNA and protein were extracted for analysis 48 h post-transfection. (C) EZH2 overexpression does not affect CBX6 expression after knockdown of EED, as detected by qRT-PCR with GAPDH for normalization. Data are presented as means ± S.D. of three independent experiments performed in triplicate. *P < 0.05. MCF-7 cells were transfected with control siRNA or siEED for 24 h, followed by transient transfection of EZH2 or empty vector for 24 h. Total RNA was then extracted for analysis. (D) Enrichment of H3K27me3 at CBX6 promoter as detected by ChIP-qPCR, *P < 0.05.
CBX6 inhibits proliferation and induces cell cycle arrest in MCF-7 cells
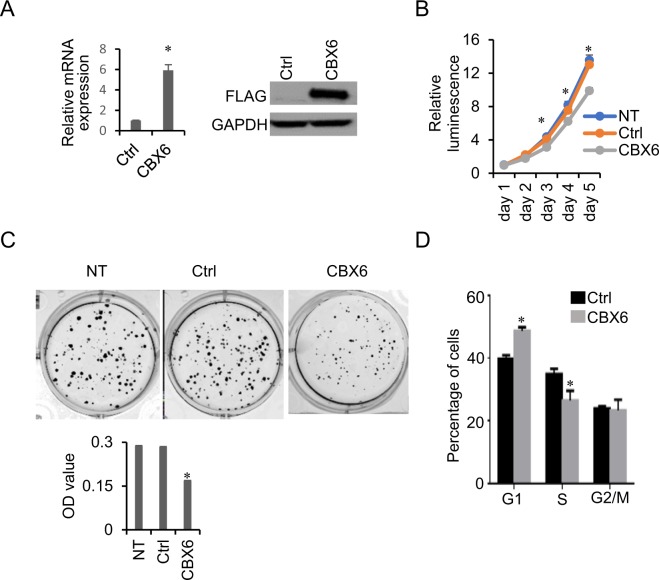
To determine the biological significance of CBX6 dysregulation, we generated stable MCF-7 cell lines expressing vector alone or FLAG-tagged CBX6 (Fig. 4A) and examined the effects of CBX6 overexpression on cell growth. As shown in Fig. 4B, CBX6-overexpressing MCF-7 cells grew moderately slower, compared with control cells transfected with the empty vector. Moreover, CBX6-overexpressing cells formed fewer and smaller colonies relative to the control group (empty vector) (Fig. 4C upper panel). This effect was further confirmed by measuring absorbance after solubilization of cell-bound crystal violet (Fig. 4C, lower panel). To assess whether the lower growth rate of CBX6-overexpressing cells is attributable to alterations in the cell cycle, we examined cell cycle distribution via flow cytometry. Our data showed a higher percentage of CBX6-overexpressing cells in the G1 phase and a lower percentage in the S phase, compared with control cells expressing empty vector (Fig. 4D).
Figure 4.
CBX6 inhibits cell proliferation and induces cell cycle arrest in MCF-7 cells. (A) Stable overexpression of CBX6-FLAG in MCF-7 cells. MCF-7 cells were transfected with CBX6-FLAG vector, then subjected to 1 mg/mL G418 selection to obtain MCF-7 cells stably overexpressing CBX6. qRT-PCR and western blot analysis were conducted to confirm the overexpression of CBX6-FLAG in the stable cell line. MCF-7 cells were transfected with an empty vector to serve as a control (Ctrl). (B) Non-transfected (NT) MCF-7 cells, MCF-7 cells stably transfected with the empty vector (Ctrl), or CBX6-FLAG (CBX6) were seeded into 96-well plates. Cell proliferation was assessed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) for 5 days. A representative of three independent experiments performed in triplicate is shown. *P < 0.05. (C) Colony formation assay. Non-transfected (NT) MCF-7 cells, MCF-7 cells stably transfected with the empty vector (Ctrl), or CBX6-FLAG (CBX6) were seeded into 6-well plates for 15 days. Colonies were stained with 0.1% crystal violet and methanol was added to solubilize the dye. Optical density (OD) values of absorbance at 540 nm was measured. Data are representative of three independent experiments performed in triplicate. *P < 0.05. (D) CBX6 induces cell cycle arrest. Control (empty vector) and CBX6-overexpressing MCF-7 cells were seeded into 6-well plates and synchronized via starvation (without serum) for 24 h. Cells were then cultured under normal conditions for 24 h and analyzed by flow cytometry. Data are representative of three independent experiments performed in triplicate, *P < 0.05.
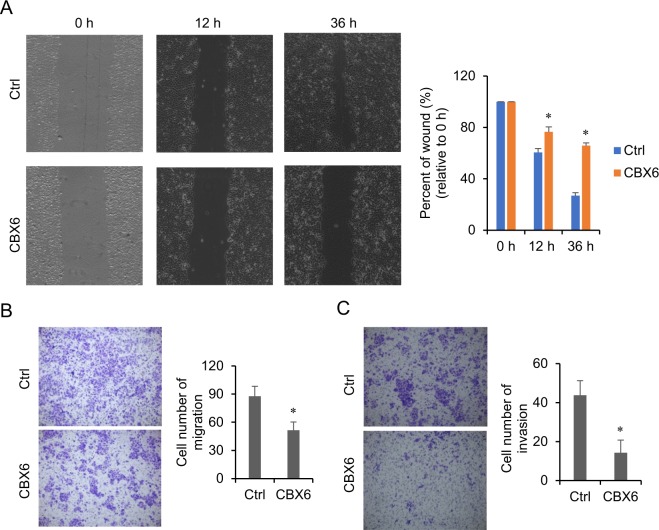
CBX6 inhibits migration and invasion of MCF-7 cells
Metastasis is an essential hallmark of cancer. To investigate the effects of CBX6 on metastasis and invasion, we performed cell migration and invasion assays in vitro. In the wound healing assay, CBX6-overexpressing MCF-7 cells showed delayed wound healing closure, compared to control cells (Fig. 5A). Similarly, CBX6 overexpression induced a dramatic reduction in migration and invasion of MCF-7 cells in a Transwell assay (Fig. 5B,C).
Figure 5.
CBX6 inhibits migration and invasion of MCF-7 cells. (A) Wound healing assay. The wound gaps were generated after cells reached 90% confluence. Images of cells were obtained at 0, 12, and 36 h post-wounding with the EVOS FL cell imaging system (Invitrogen). Representative images of the wound healing assay were shown on the left. The wound area was measured, and percent of initial wound area was compared between MCF-7 cells stably transfected with the empty vector (Ctrl), or CBX6-FLAG (CBX6) (right panel). *P < 0.05. (B,C) Transwell invasion and migration assay. Representative images of migrated (B) or invaded (C) cells were shown on the left. Cell numbers were quantified by counting ten random fields at ×200 magnification. MCF-7 cells stably transfected with CBX6-FLAG (CBX6) have decreased abilities of migration and invasion, compared to the cells stably transfected with the empty vector (Ctrl). *P < 0.05.
Gene expression changes induced by CBX6 overexpression
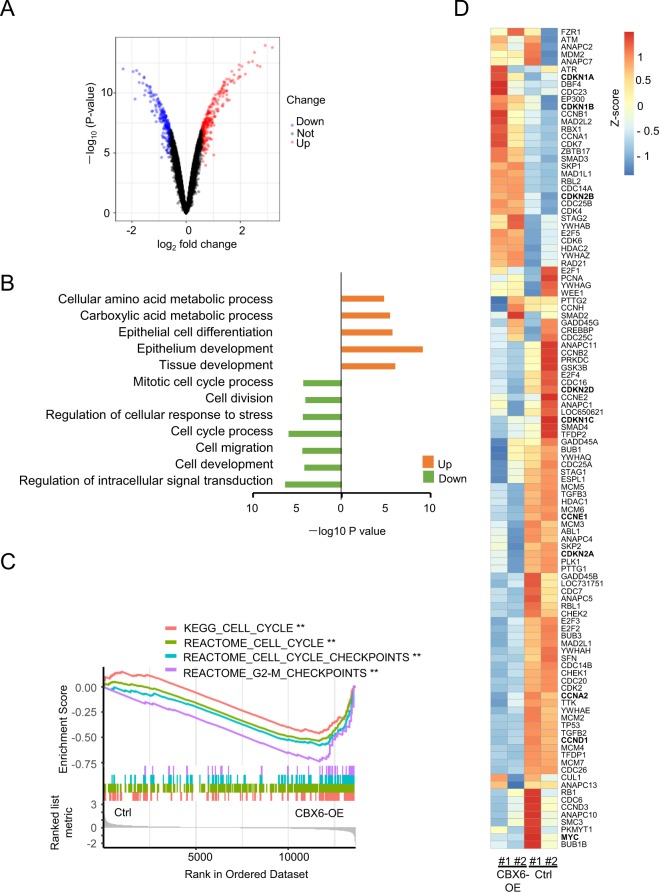
To clarify the mechanisms underlying CBX6 activity in breast cancer progression, MCF-7 cells stably overexpressing the gene were subjected to microarray analysis. In total, 525 genes were differentially expressed between CBX6-overexpressing and vector control cells, among which 234 were downregulated (log2 fold change <−0.6, P < 0.05) and 291 were upregulated (log2 fold change >0.6, P < 0.05) in CBX6-overexpressing cells (Fig. 6A; Supplementary Table S2). Gene ontology analysis revealed that upregulated genes in CBX6-overexpressing cells are involved in epithelial cell differentiation and epithelium development whereas downregulated genes participate in the cell cycle, cell division, cell migration, cell development, regulation of intracellular signal transduction and cellular response to stress (Fig. 6B).
Figure 6.
Effects of CBX6 overexpression on gene expression in MCF-7 cells. (A) Gene expression was examined by HumanHT-12 v4Expression BeadChip microarrays (Illumina). The volcano plot shows statistical significance (−log10 (P-value) plotted against log2 fold change of genes for CBX6-overexpressing cells against control (empty vector) cells. Differentially expressed genes were selected based on criteria of P < 0.05 and absolute log2 fold change >0.6. (B) Gene ontology (GO) analysis of significantly altered genes in CBX6-overexpressing cells. (C) Gene set enrichment analysis (GSEA) of gene expression profiles of MCF-7 cells overexpressing CBX6 (CBX6-OE) versus control. Shown are results using REACTOME “Cell Cycle”, “Cell Cycle Checkpoints”, “G2-M Checkpoints” and Kyoto Encyclopedia of Genes and Genomes (KEGG) “Cell Cycle” as the interrogating gene sets. **P ≤ 0.01. (D) The expression heatmap of genes included in the gene set of KEGG “Cell Cycle” in MCF-7 cells overexpressing CBX6 (CBX6-OE), and control MCF-7 cells (Ctrl) which are stably transfected with the empty vector.
To gain more insights into the functional consequences of restoring CBX6 expression in MCF7 cells, we performed gene set enrichment analysis (GSEA), and found that multiple cell cycle genes, included in the gene matrices of the Kyoto Encyclopedia of Genes and Genomes (KEGG) Cell cycle, REACTOME cell cycle, REACTOME cell cycle checkpoints and REACTOME G2_M checkpoints, are downregulated in CBX6-overexpressing (CBX6-OE) MCF-7 cells (Fig. 6C). Specifically, several cyclins including G1/S Cyclin CCND1 and CCNE1 are downregulated in CBX6-OE MCF-7 cells, consistent with the slight G1-arrest in CBX6-OE cells. Meanwhile, the cyclin-dependent kinase inhibitor gene, including CDKN1B (p27), CDKN1A (p21) are upregulated in CBX6-OE MCF-7 cells. CDKN2A (p16INK4A/p14ARF) is a known Polycomb target gene32; its expression is downregulated in CBX6-OE MCF-7 cells. Paradoxically, CDK2B, the neighboring gene of CDKN2A, is upregulated in CBX6-OE MCF-7 cells. MYC, the oncogene which amplifies the output of the existing gene expression program33, and promotes tumorigenesis by regulating cell growth, proliferation and metabolism34, is downregulated by CBX6 overexpression (Fig. 6D). All these results indicate that CBX6 governs a complex gene expression program.
BST2 is directly downregulated by CBX6
PcG proteins have context-dependent actions on gene expression35. We reasoned high-confidence direct targets of CBX6 should have a greater tendency to be modulated by CBX6 in different cell line models. To ascertain potentially significant genes in the CBX6-associated tumorigenesis, we performed microarray analysis on a U251MG glioma cell line overexpressing CBX6 that we previously established (Deng et al., unpublished results). Searching for genes affected led to the identification of multiple genes repressed by CBX6’s overexpression in both cell lines, including bone marrow stromal cell antigen 2 (BST2). BST2, which encodes a type II integral membrane protein that inhibits the release of enveloped viruses through its homodimerization36,37, is overexpressed in multiple cancers including breast cancer37. BST2 was demonstrated to promote tumor survival, invasion and metastasis through a plethora of mechanisms37,38.
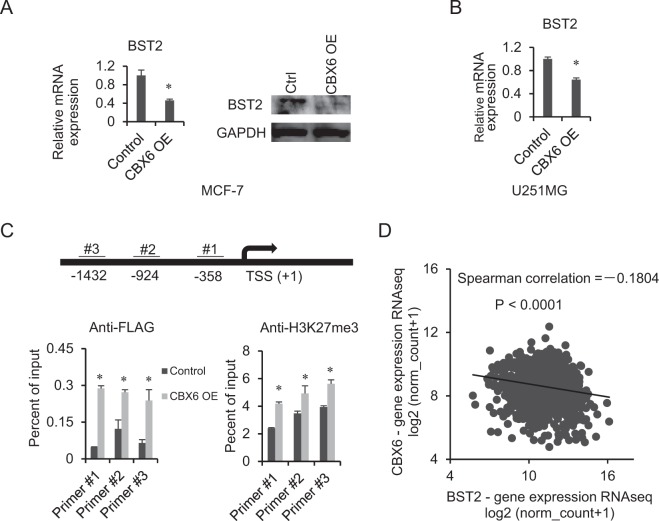
We applied qRT-PCR and western blot analyses to confirm the regulation of BST2 by CBX6. As expected, overexpression of CBX6 in MCF-7 cells was associated with decreased levels of BST2 mRNA and protein (Fig. 7A). Examination of the CBX6-OE U251MG cells and its control revealed that overexpression of CBX6 also caused a significant decrease in BST2 in U251MG cells (Fig. 7B). We further determined whether CBX6 directly targets BST2. As shown in Fig. 7C, both CBX6 and H3K27me3 were significantly enriched at the BST2 promoter in CBX6-OE MCF-7 cells. Moreover, examination of TCGA dataset disclosed a marked increase in BST2 expression in breast cancer (Supplementary Fig. S5) and CBX6 and BST2 were negatively correlated in breast cancer (Spearman r = −0.1804, P < 0.0001) (Fig. 7D). Based on these findings, we propose that CBX6 potentially plays a tumor suppressor role through repression of BST2 in breast cancer.
Figure 7.
CBX6 directly downregulates the expression of Bone Marrow Stromal cell antigen 2 (BST2). (A) CBX6 downregulates BST2 expression in MCF-7 cells. Left: qRT-PCR analysis of BST2 mRNA in the control (empty vector) and CBX6-overexpressing (CBX6-OE) cells using GAPDH for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. Right: Western blot analysis of BST2 in control (empty vector) and CBX6-overexpressing cells (CBX6-OE) using GAPDH as a loading control. (B) CBX6 downregulates BST2 expression in U251MG cells. qRT-PCR analysis of BST2 mRNA levels in control (empty vector) and CBX6-overexpressing cells using GAPDH for normalization. Data are presented as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. (C) Enrichment of CBX6 and H3K27me3 at the BST2 promoter detected via ChIP-qPCR. Top: The locations of primers at the BST2 promoter. Bottom: Enrichment of CBX6 and H3K27me3 at the BST2 promoter in control (empty vector) and CBX6-overexpressing cells. Data are presented as a percentage of the input, and as means ± S.D. from three independent experiments performed in triplicate. *P < 0.05. (D) Spearman’s rank correlation coefficient indicating a negative correlation between CBX6 and BST2 mRNA levels in breast cancer tissues based on The Cancer Genome Atlas (TCGA) RNA-Seq data.
Discussion
PcG-mediated gene silencing plays a critical role in cancer development
CBX6, a subunit of PRC1, reads the crucial epigenetic marker, H3K27me3, written by PRC2. Recent studies have reported that CBX6 assembles into PRC1 and its knockdown via shRNA induces spontaneous differentiation of embryonic stem cells (ESC), supporting a role as an essential regulator of ESC identity39. Ectopic expression of CBX6 is reported to block reprogramming40 and its dysregulation implicated in different types of malignancies23,24. Experiments in the current study revealed significant downregulation of CBX6 in breast cancer. CBX6 inhibited cell proliferation and induced cell cycle arrest in MCF-7 in vitro. Moreover, cell migration and invasion rates were significantly reduced upon CXB6 overexpression.
We examined changes in gene expression profiles caused by CBX6 overexpression via microarray analysis. Consistent with functional experimental results, genes downregulated in association with CBX6 overexpression are involved in the cell cycle, division and migration. For example, CBX6 represses expressions of G1/S Cyclin CCND1 and CCNE1 in MCF-7 cells. Both CCND1 and CCNE1 were reported to be repressed by PcG proteins: CBX7, a paralog of CBX6, binds the CCNE1 prompter and represses the expression of CCNE1 partly through HDAC2 in mice and human lung carcinomas and gliomas41,42; JARID2, a co-factor facilitating genomic targeting of PRC2, represses CCND1’s expression in cardiomyocytes and leukemia43–45. In this study, we also found CBX6 repressed MYC expression in MCF-7 cells. Kaur et al. reported that MYC limits its own expression by a feedback loop mediated by EZH246,47, the finding here suggests CBX6 might participate in the process. In addition to amplifying transcription, paradoxically, MYC represses discrete sets of genes through direct and indirect mechanisms48. Best characterized MYC-repressed genes include CDKN1A, CDKN1B and CDKN2B48, all of which are upregulated in the MCF-7 cell upon CBX6 over-expression, suggesting a potential CBX6-MYC-CDK inhibitors pathway regulating cell cycle progression. Nevertheless, the results here indicate that CBX6 regulates complex regulatory circuits, further research is needed to ascertain the effects of CB6 on gene expression, elucidate the detailed mechanisms by which CBX6 controls gene expression, directly or indirectly.
In addition to breast cancer, CBX6 is reported to be downregulated in glioblastomas although the underlying mechanisms are still unknown. EZH2 is highly expressed in multiple cancers, including glioblastoma and breast cancer. Moreover, high expression of EZH2 is associated with aggressive disease and poor outcome49–51. To date, several small molecule inhibitors targeting EZH2 have been investigated in preclinical and clinical trials52. PcG proteins are suggested to auto-regulate their own expression in human embryonic fibroblasts53. Overexpression and knockdown experiments in the current study showed that EZH2, the catalytic subunit of PRC2, repressed the expression of CBX6. Earlier studies suggest that gene expression regulated by EZH2 is either PRC2-dependent or independent54,55. Here, we observed upregulation of CBX6 expression by the EZH2 inhibitor EPZ-6438. Knockdown of EED led to upregulation of CBX6 in MCF-7 cells. Additionally, EZH2 overexpression did not affect CBX6 expression after knockdown of EED in MCF-7 cells. Our data collectively indicate that downregulation of CBX6 by EZH2 requires intact PRC2 activity.
Bone marrow stromal cell antigen 2 (BST2), also known as CD317 or tetherin, is a lipid raft-associated type 2 transmembrane glycoprotein. BST2 promotes activation of NF-kB, leading to the production of proinflammatory factors involved in inhibition of viral replication56,57 and additionally implicated in blocking virion release via physically tethering to the cell surface58. BST2 may be involved in cell-to-cell interactions in view of the finding that purified extracellular domains of the protein inhibit the adhesion of human monocytes to HUVECs59. Inflammatory responses and cell adhesion contribute to tumorigenesis, indicating a role in cancer progression. Accumulating studies have provided evidence that BST2 is involved in tumor progression in many cancer types, including breast cancer60–67. Yi et al. demonstrated that BST2 is upregulated in tamoxifen-resistant breast cancer cells and enhances the invasion and migration capacities of tumor cells. Mahauad-Fernandez’s research group reported that knockdown of BST2 inhibits mammary tumor growth and metastasis, both in vitro and in vivo, and high BST2 expression in breast tumors is positively associated with tumor size and aggressiveness as well as poor patient survival63. In addition, monoclonal antibodies against BST2 possess significant antitumor activity in lymphoma and endometrial cancers68,69. In view of the combined results, BST2 is proposed as an ideal therapeutic target for breast cancer. In the current study, CBX6 significantly suppressed BST2 expression in both MCF-7 and U251MG cells, implying that this process is independent of cell type. Additionally, CBX6 bound directly to the BST2 promoter and increased enrichment of H3K27me3.
In conclusion, our experiments provide evidence of significant downregulation of CBX6 expression in breast cancer. CBX6 was negatively regulated by EZH2 in a PRC2-dependent manner. Overexpression of CBX6 inhibited the proliferation and metastasis capacity in breast cancer cells in vitro, altered the expression of genes involved in cell cycle regulation and other pathways. Moreover, CBX6 exerted a marked suppressive effect on expression of BST2 through binding to its promoter and altering histone modification. To our knowledge, this is the first study to demonstrate a tumor suppressor role of CBX6 in breast cancer.
Methods
Bioinformatic analysis
mRNA expression profiles and clinical information on the 1119 breast carcinomas and 113 normal samples were obtained from The Cancer Genome Atlas (TCGA) data portal. Gene set enrichment analysis70 was performed as described71. Kaplan-Meier survival analyses were performed using GraphPad Prism 6 or the Kaplan-Meier plotter (www.kmplot.com)29.
Cell culture and drug treatment
MCF-7 and MDA-MB-231 were kindly provided by Professor Edwin Chong Wing Cheung (Faculty of Health Sciences, University of Macau) and MCF-10A by Professor Lijun Di (Faculty of Health Sciences, University of Macau). C2C12 was purchased from the Cell Resources Center of Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). MCF-7, MDA-MB-231 and C2C12 cells were cultured in DMEM (Gibco) containing 10% FBS. MCF10A cells were cultured in DMEM supplemented with 10% FBS, 0.5 µg/mL hydrocortisone (Sigma), 10 µg/mL insulin (Thermo Fisher), and 20 ng/mL hEGF (Thermo Fisher). All cells were tested negative for mycoplasma contamination and were maintained at 37 °Cand 5% CO2. For drug treatment, MCF-7 cells were seeded in 6-well plates and incubated overnight, followed by treatment with 50 µm EPZ6438 (Selleck Chemicals). Control cells were treated with 0.1% DMSO. All cells were treated for 5 consecutive days followed by extraction of protein and RNA for analysis.
Plasmids and transfection
Human CBX6-Myc-DDK cloned into pCMV6_Entry was purchased from OriGene (RC204166). Human EZH2 was amplified using cDNA derived from MCF-7 cells as a template and cloned into pcDNA3.2/GW/D-TOPO expression vector (Invitrogen). Expression vectors were transfected into MCF-7 cells with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Cells transfected with the corresponding empty expression vector served as the controls. At 48 h post-transfection, protein and RNA were isolated for analysis. For generating a CBX6-expressing stable cell line, 1 mg/mL G418 (Gibco) was added to the culture medium to screen for stable clones. Expression of CBX6 mRNA and protein was verified via qRT-PCR and western blot analysis, respectively. For knockdown experiments, a pool of siRNAs targeting EZH2 (Santa Cruz Biotechnology; sc35312) and three EED siRNAs (Integrated DNA Technologies (IDT)) were employed. MCF-7 cells were transfected with test or control siRNA at a final concentration of 50 nM using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s instructions. At 48 h post-transfection, protein and RNA were harvested for analysis.
Cell proliferation assay
MCF-7 cells (2 × 103) were plated in 96-well plates in 100 µL medium. Cell proliferation was analyzed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) for five consecutive days.
Colony formation assay
MCF-7 cells were seeded into 6-well plates at a density of 500 cells per well. At 15 days after seeding, each well was stained with 0.1% crystal violet and methanol added to solubilize the dye. Absorbance at 540 nm was read using a Spark Multimode microplate reader (Tecan).
Flow cytometry analysis of the cell cycle
MCF-7 cells were seeded into 6-well plates and synchronized via starvation (without serum) for 24 h, followed by culturing under normal conditions for 24 h. Cells were harvested and stained using cell cycle and apoptosis analysis kits (Beyotime) and analyzed with a FACS scan flow cytometer (BD Biosciences). The relative ratios of G1, S, and G2 phases were analyzed using the FlowJo 2.8 software.
Wound healing assay
MCF-7 cells were seeded into 6 well-plates. After cells reached 90% confluence, cross lines were generated using a sterile pipette tip. Images of cells were obtained after 0, 12 and 36 h post-wounding using the EVOS FL cell imaging system (Thermo Fisher).
Transwell assay
The Transwell assay was performed in 24-well chambers (Corning). For the migration assay, 2.5 × 105 cells were resuspended in serum-free medium and added to the upper chamber. The bottom chamber was filled with 500 μL culture medium containing 10% FBS. After culturing for 24 hours, cells on the lower surface of the membrane were stained with 0.1% crystal violet and photographed using the Carl Zeiss Axio Observer microscope. For the invasion assay, the upper chamber surface of the basement membrane of the Transwell was coated with Matrigel (Corning). Aliquots of cells (1 × 106) were resuspended in serum-free medium and added to the upper chamber. The bottom chamber was filled with 500 μL culture medium containing 10% FBS. After culturing for 36 hours, cells on the lower surface of the membrane were stained with 0.1% crystal violet and photographed using the Carl Zeiss Axio Observer microscope.
Total RNA extraction
Total RNA was extracted using the TRIzol reagent (Invitrogen). Genomic DNA was digested with an RNase-free DNase kit (Qiagen) and subsequently purified using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. The concentration and quality of RNA were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies).
Gene expression profiling and analysis
Total RNA was amplified and labeled using the TargetAmp™-Nano Labeling Kit for Illumina Expression BeadChip (Epicentre Biotechnologies). Labeled cRNA was purified with the RNeasy mini kit (Qiagen) and hybridized on HumanHT-12 v4 Expression BeadChip microarrays (Illumina) according to the manufacturer’s protocol. Hybridized arrays were scanned using the Illumina iScan, and the image data was extracted using the Illumina Genome Studio software. Gene ontology analysis was conducted using the PANTHER Overrepresentation Test (http://www.geneontology.org/).
Quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR)
Total RNA (1 μg) was used to synthesize cDNA using a PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara). Quantitative PCR (qPCR) was conducted using the iTaq™ Universal SYBR® Green Supermix (Bio-Rad) in the CFX 96 thermocycler (Bio-Rad). Assays were performed in triplicate and repeated three times. The relative expression level of the gene of interest was normalized to that of GAPDH and calculated according to the 2−ddCt method. The primers used in this study are presented in Supplementary Table S1.
Western blot
Whole cell lysates were harvested using RIPA Lysis and Extraction Buffer (Invitrogen) supplemented with the protease inhibitor cocktail (Sigma). The extracted protein was separated via SDS-PAGE and transferred to nitrocellulose membranes (Pall Corporation). Membranes were blocked with 5% non-fat milk and incubated with the following antibodies: CBX6 (Millipore 09-030), FLAG (Sigma-Aldrich F1804), EZH2 (Cell Signaling Technology 5246 S), H3K27me3 (Abcam ab6002), EED (Millipore 05-1320), BST2 (Santa Cruz Biotechnology sc-390719), GAPDH (Cell Signaling Technology 5174 s), H3 (Abcam ab1791), followed by horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, 1:5000) for 60 min at room temperature. Signals were detected with the enhanced chemiluminescence (ECL) detection system (Thermo Fisher Scientific).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed using the EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (Millipore) in accordance with the manufacturer’s instructions. Briefly, cells were cross-linked with 1% paraformaldehyde and sonicated to obtain fragments between 0.2 and 0.5 kb. Chromatin was incubated with anti-FLAG (Sigma-Aldrich F1804), anti-H3K27me3 (Millipore 07-449) or control mouse IgG (Cell Signal Technology 5415) antibodies and protein A/G beads overnight. DNA was extracted and used for ChIP-qPCR analysis. Enrichment levels are presented as a percentage of input chromatin. The primers are presented in Supplementary Table S1.
Electronic supplementary material
Acknowledgements
This work was supported by the Science and Technology Development Fund of Macau [137/2014/A3, 095/2015/A3] and the Research & Development Administration Office of the University of Macau [SRG201400015, MYRG201500232, MYRG201700099] awarded to G.L.
Author Contributions
G.L. and H.D. conceived of the study, designed the experiments. H.D., X.G. and L.G. carried out the experiments. H.D., J.Z. and G.L. analyzed data. H.Z. and M.Y.C. contributed materials. H.D. and G.L. drafted the manuscript. All authors have read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36560-4.
References
- 1.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Molecular cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Cai L, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Molecular cell. 2013;49:571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Current opinion in genetics & development. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endoh M, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS genetics. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, N.Y.) 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Y, et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nature communications. 2017;8:672. doi: 10.1038/s41467-017-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem cells (Dayton, Ohio) 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- 12.Entrevan M, Schuettengruber B, Cavalli G. Regulation of Genome Architecture and Function by Polycomb Proteins. Trends in cell biology. 2016;26:511–525. doi: 10.1016/j.tcb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Vizan P, Beringer M, Ballare C, Di Croce L. Role of PRC2-associated factors in stem cells and disease. The FEBS journal. 2015;282:1723–1735. doi: 10.1111/febs.13083. [DOI] [PubMed] [Google Scholar]
- 14.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell death & disease. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Nigris F. Epigenetic regulators: Polycomb-miRNA circuits in cancer. Biochimica et biophysica acta. 2016;1859:697–704. doi: 10.1016/j.bbagrm.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Kaustov L, et al. Recognition and specificity determinants of the human cbx chromodomains. The Journal of biological chemistry. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont PL, et al. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. British journal of cancer. 2014;111:1663–1672. doi: 10.1038/bjc.2014.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scales M, Jager R, Migliorini G, Houlston RS, Henrion M. Y. visPIG–a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PloS one. 2014;9:e107497. doi: 10.1371/journal.pone.0107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer research. 2016;76:7277–7289. doi: 10.1158/0008-5472.can-16-2100. [DOI] [PubMed] [Google Scholar]
- 20.Pallante P, Forzati F, Federico A, Arra C, Fusco A. Polycomb protein family member CBX7 plays a critical role in cancer progression. American journal of cancer research. 2015;5:1594–1601. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, C. Z. et al. CBX8 exhibits oncogenic activity via AKT/beta-Catenin activation in hepatocellular carcinoma. Cancer research, 10.1158/0008-5472.can-17-0700 (2017). [DOI] [PubMed]
- 22.Wang G, et al. CBX8 Suppresses Tumor Metastasis via Repressing Snail in Esophageal Squamous Cell Carcinoma. Theranostics. 2017;7:3478–3488. doi: 10.7150/thno.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PloS one. 2013;8:e80970. doi: 10.1371/journal.pone.0080970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei S, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic acids research. 2017;45:D658–d662. doi: 10.1093/nar/gkw983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen WY, et al. Chromobox homolog 2 protein: A novel biomarker for predicting prognosis and Taxol sensitivity in patients with breast cancer. Oncology letters. 2017;13:1149–1156. doi: 10.3892/ol.2016.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng JS, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. The international journal of biochemistry & cell biology. 2017 doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Kim HY, Park JH, Won HY, Lee JY, Kong G. CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/beta-catenin pathway. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015;29:300–313. doi: 10.1096/fj.14-253997. [DOI] [PubMed] [Google Scholar]
- 28.Chung CY, et al. Cbx8 Acts Non-canonically with Wdr5 to Promote Mammary Tumorigenesis. Cell reports. 2016;16:472–486. doi: 10.1016/j.celrep.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 30.Liang YK, Lin HY, Chen CF. & Zeng. Prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8:92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. Clinical and prognostic relevance of EZH2 in breast cancer: A meta-analysis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;75:218–225. doi: 10.1016/j.biopha.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 32.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature reviews. Molecular cell biology. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 33.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppens M, van Lohuizen M. Context-dependent actions of Polycomb repressors in cancer. Oncogene. 2016;35:1341–1352. doi: 10.1038/onc.2015.195. [DOI] [PubMed] [Google Scholar]
- 36.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nature immunology. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahauad-Fernandez WD, Okeoma CM. The role of BST-2/Tetherin in host protection and disease manifestation. Immunity, inflammation and disease. 2016;4:4–23. doi: 10.1002/iid3.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter-Yohrling J, et al. Identification of genes expressed in malignant cells that promote invasion. Cancer research. 2003;63:8939–8947. [PubMed] [Google Scholar]
- 39.Santanach A, et al. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nature communications. 2017;8:1235. doi: 10.1038/s41467-017-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ning B, et al. USP26 functions as a negative regulator of cellular reprogramming by stabilising PRC1 complex components. Nature communications. 2017;8:349. doi: 10.1038/s41467-017-00301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forzati F, et al. CBX7 is a tumor suppressor in mice and humans. The Journal of clinical investigation. 2012;122:612–623. doi: 10.1172/jci58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu T, et al. CBX7 is a glioma prognostic marker and induces G1/S arrest via the silencing of CCNE1. Oncotarget. 2017;8:26637–26647. doi: 10.18632/oncotarget.15789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyoda M, et al. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Developmental cell. 2003;5:85–97. doi: 10.1016/S1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- 44.Nakajima K, et al. Coordinated regulation of differentiation and proliferation of embryonic cardiomyocytes by a jumonji (Jarid2)-cyclin D1 pathway. Development (Cambridge, England) 2011;138:1771–1782. doi: 10.1242/dev.059295. [DOI] [PubMed] [Google Scholar]
- 45.Su CL, Deng TR, Shang Z, Xiao Y. JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. International journal of hematology. 2015;102:76–85. doi: 10.1007/s12185-015-1797-x. [DOI] [PubMed] [Google Scholar]
- 46.Kaur, M. & Cole, M. MYC acts via the PTEN tumor suppressor to elicit autoregulation and genome-wide gene repression by activation of the Ezh2 methyltransferase. Cancer research, 10.1158/0008-5472.Can-12-2522 (2012). [DOI] [PMC free article] [PubMed]
- 47.Cole MD. MYC association with cancer risk and a new model of MYC-mediated repression. Cold Spring Harbor perspectives in medicine. 2014;4:a014316. doi: 10.1101/cshperspect.a014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nature reviews. Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 49.Beca F, et al. EZH2 protein expression in normal breast epithelium and risk of breast cancer: results from the Nurses’ Health Studies. Breast cancer research: BCR. 2017;19:21. doi: 10.1186/s13058-017-0817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song X, et al. Selective inhibition of EZH2 by ZLD1039 blocks H3K27 methylation and leads to potent anti-tumor activity in breast cancer. Scientific reports. 2016;6:20864. doi: 10.1038/srep20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, et al. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer letters. 2015;356:929–936. doi: 10.1016/j.canlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nature medicine. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, N.Y.) 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahara S, et al. HIFI-alpha activation underlies a functional switch in the paradoxical role of Ezh2/PRC2 in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E3735–3744. doi: 10.1073/pnas.1602079113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokarev A, et al. Stimulation of NF-kappaB activity by the HIV restriction factor BST2. Journal of virology. 2013;87:2046–2057. doi: 10.1128/jvi.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell host & microbe. 2012;12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 59.Yoo H, Park SH, Ye SK, Kim M. IFN-gamma-induced BST2 mediates monocyte adhesion to human endothelial cells. Cellular immunology. 2011;267:23–29. doi: 10.1016/j.cellimm.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Kuang CM, et al. BST2 confers cisplatin resistance via NF-kappaB signaling in nasopharyngeal cancer. Cell death & disease. 2017;8:e2874. doi: 10.1038/cddis.2017.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukai S, et al. Overexpression of Transmembrane Protein BST2 is Associated with Poor Survival of Patients with Esophageal, Gastric, or Colorectal Cancer. Annals of surgical oncology. 2017;24:594–602. doi: 10.1245/s10434-016-5100-z. [DOI] [PubMed] [Google Scholar]
- 62.Mahauad-Fernandez WD, Borcherding NC, Zhang W, Okeoma CM. Bone marrow stromal antigen 2 (BST-2) DNA is demethylated in breast tumors and breast cancer cells. PloS one. 2015;10:e0123931. doi: 10.1371/journal.pone.0123931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahauad-Fernandez WD, DeMali KA, Olivier AK, Okeoma CM. Bone marrow stromal antigen 2 expressed in cancer cells promotes mammary tumor growth and metastasis. Breast cancer research: BCR. 2014;16:493. doi: 10.1186/s13058-014-0493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sayeed A, et al. Aberrant regulation of the BST2 (Tetherin) promoter enhances cell proliferation and apoptosis evasion in high grade breast cancer cells. PloS one. 2013;8:e67191. doi: 10.1371/journal.pone.0067191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones PH, Mahauad-Fernandez WD, Madison MN, Okeoma CM. BST-2/tetherin is overexpressed in mammary gland and tumor tissues in MMTV-induced mammary cancer. Virology. 2013;444:124–139. doi: 10.1016/j.virol.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi EH, et al. BST-2 is a potential activator of invasion and migration in tamoxifen-resistant breast cancer cells. Biochemical and biophysical research communications. 2013;435:685–690. doi: 10.1016/j.bbrc.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 67.Cai D, et al. Up-regulation of bone marrow stromal protein 2 (BST2) in breast cancer with bone metastasis. BMC cancer. 2009;9:102. doi: 10.1186/1471-2407-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schliemann C, et al. In vivo biotinylation of the vasculature in B-cell lymphoma identifies BST-2 as a target for antibody-based therapy. Blood. 2010;115:736–744. doi: 10.1182/blood-2009-08-239004. [DOI] [PubMed] [Google Scholar]
- 69.Yokoyama T, et al. Plasma membrane proteomics identifies bone marrow stromal antigen 2 as a potential therapeutic target in endometrial cancer. International journal of cancer. 2013;132:472–484. doi: 10.1002/ijc.27679. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng H, et al. Histone H3.3K27M Mobilizes Multiple Cancer/Testis (CT) Antigens in Pediatric Glioma. Molecular cancer research: MCR. 2018;16:623–633. doi: 10.1158/1541-7786.Mcr-17-0460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.