Abstract
An important factor in quality control of non-invasive prenatal screening (NIPS) or testing (NIPT) is a sufficient percentage of fetal DNA to avoid false-negative results. Here we evaluate 14,379 shallow whole-genome sequenced diagnostic NIPS samples, as well as negative controls, for both technical and biological factors that can influence fetal fraction and its assessment. Technically, bioinformatics analyses can have a profound impact on fetal fraction determination. We found best performance for fetal fraction determination with the Y chromosome based tool DEFRAG for male fetuses and the count based tool SeqFF for female fetuses. Biologically, gestational age of up to 21 weeks and maternal age had no influence on fetal fraction, while an increase in weight and BMI had a negative influence on fetal fraction. While a trend was observed, no statistically significant difference in fetal fraction was found between trisomy and normal samples. Overall, these results confirm the influence of biological factors and give insight into technical factors that can affect fetal fractions in NIPS.
Subject terms: High-throughput screening, Genetics research
Introduction
Non-invasive prenatal screening (NIPS) or testing (NIPT) for aneuploidy detection should be evaluated for sufficient fetal DNA fraction (FF) since low levels may give rise to false-negative results. Indeed, some laboratories may not be checking FF or using optimal methods for detection that could potentially provide false-negative results to patients [1]. Technical factors that can influence FF and its estimation are sample handling [2] and the choice of bioinformatics tools (reviewed in [3]) [4]. Multiple biological factors have been identified that affect FF, including gestational age [4–9], weight and/or body mass index (BMI) [6–12], trisomies [7, 10–13], fetal crown-rump length [6, 10, 12], serum pregnancy-associated plasma protein-A [6, 10, 12], serum free β-human chorionic gonadotropin [6, 10, 12], hypertension [7], twins [7], smoking [10], and assisted conception [8, 12]. Here we evaluate technical and biological variables influencing FF and its determination in a diagnostic setting.
Subjects and Methods
NIPS was successfully performed on 14,379 samples with consent for research, from 01 April 2017 to 01 October 2017, at the VU University Medical Center Amsterdam as an expansion of the nationwide TRIDENT [14, 15] study, including high and low-risk pregnancies. Ten non-pregnant women also provided blood to serve as negative controls.
Blood was shipped in Streck tubes (La Vista, NE, USA: 10 ml Cell-Free DNA BCT CE) and shallow whole-genome sequenced (Supplemental File 1). Reads were then aligned and evaluated for aneuploidies with WISECONDOR [16]. To simulate variable FF, a titration series was generated by pairing 10 negative controls (non-pregnant females) and 10 randomly selected male fetus samples and mixing reads from the pairs at increments of ten percent (Supplemental File 1). FF for the titration and diagnostic samples were determined with DEFRAG [4] (whole and subset of Y chromosome methods, henceforth referred to as DEFRAG_W and DEFRAG_S, respectively), SANEFALCON [17], and SeqFF [18] (including ENET [19] and WRSC [20] scores). Fetal sex was determined as a consensus gender call from DEFRAG_W and DEFRAG_S or the test was considered failed if in contradiction. For statistical analyses, bioinformatics FF results were evaluated using Spearman correlations, trisomy associations compared with logistic models, and all other FF associations using logistic models with ANOVA (based upon a chi-square test). More detailed methods are provided in Supplemental File 1.
Results and Discussion
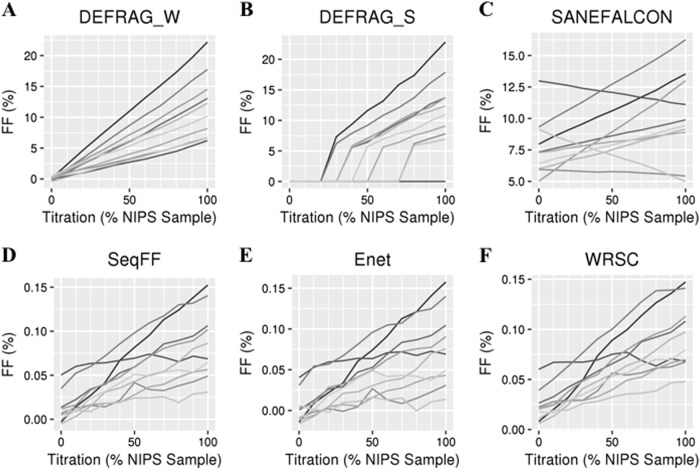
In a 6-month period, 7,397 male fetuses and 6,982 female fetuses were successfully evaluated by NIPS with a mean / median of 15,437,326 / 15,334,562 non-duplicate aligned reads (Supplemental Fig. 1). Bioinformatics tools had considerably different performances in determining FFs. In a titration series from mixing non-pregnant with pregnant samples, DEFRAG_W was the only tool showing an expected distribution of fetal DNA (Fig. 1a). Since DEFRAG_S only uses a small Y-specific bin number and also reports medians it has a trend to report no or high FF (Fig. 1b), and therefore calls low or medium levels as negative. SeqFF, Enet, and WRSC overall perform well with a trend for increasing FF at higher levels of pregnant samples (Fig. 1d-f). The poorest performing tool was SANEFALCON with some titrations having a negative slope due to the high false-positive FF calls (Fig. 1c). Based on these data, we recommend DEFRAG_W for FF determination in pregnancies with male fetuses.
Fig. 1.
FF determinations by each bioinformatics tool (a–f) based on ten synthetic titration series (each a different shade of gray) from 100% control (not pregnant, 0% on x-axis) samples to 100% NIPS samples
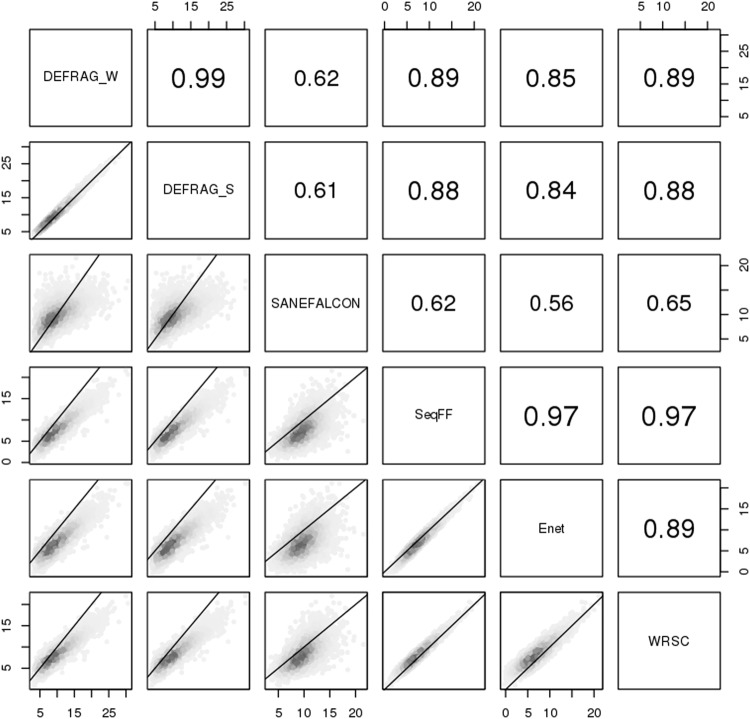
However, DEFRAG_W cannot be used on female fetuses since it is Y chromosome based. Comparing the non-Y chromosome based tools to DEFRAG_W (the best performing tool based on titration analyses), we find the highest agreement with SeqFF (Fig. 2). We do observe differences in intercepts when comparing tools (Fig. 2) and FF distributions among tools (Supplemental Fig. 2) which implies that tools with high correlation may systematically predict higher or lower FF. For example, on average SeqFF predicts a FF of 2.34% less than DEFRAG_W. Hence, minimal FF cutoffs for trisomy detection should be evaluated on a per tool basis. Overall, since SeqFF performed the best out of all non-Y chromosome based tools we recommend using it for FF determination in female fetuses.
Fig. 2.
Bioinformatic comparisons for male fetuses FF. Numbers within panels indicate spearman correlations. Axis numbers indicate FF (%). Lines indicate theoretical one-to-one values
Overall, we confirm a high correlation between DEFRAG_W and SeqFF [4], though further extrapolate that DEFRAG_W routinely gives higher predictive values than SeqFF. However, SeqFF did detect non-zero FF for two 100% negative control samples, and therefore suggest it could use further optimization. These could potentially be explained as non-pregnant women having shorter cell-free DNA than pregnant women [21]. Since DNA that is fetal in origin also tends to be shorter, non-pregnant control DNA with shorter distributions may be misinterpreted as fetal in origin. A large limitation of evaluating FF is not knowing the biological truth, other than for negative controls. Therefore, future studies should include secondary methods for FF determination to more reliably establish best bioinformatics tools and cutoffs. On the basis of these evaluations, we continued association analyses using DEFRAG_W for male fetuses FF and SeqFF for female fetuses FF.
To determine if shipping time could influence the FF in blood samples, we evaluated FF as a function of shipping time. The longest shipping time within this cohort was 1 week, with most samples (73%) taking two days or less (Supplemental Fig. 3). Within this time interval we saw no correlation between shipping time and FF (Supplemental Fig. 3), confirming previous reports [2] and fitting within manufacturer guidelines (up to 14 days at 6–37 °C) when using Streck tubes.
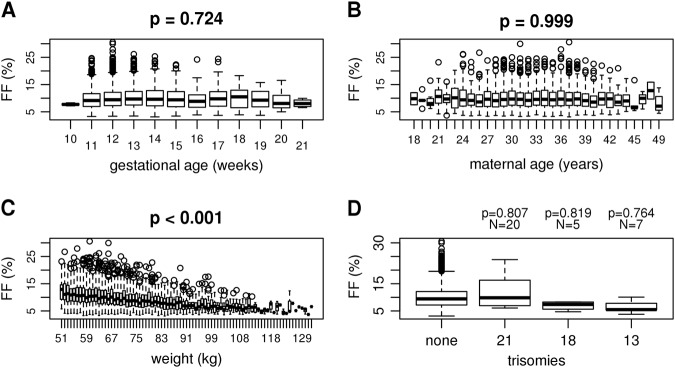
The general consensus in literature is that FF increases with later gestational age [6–9]. In our cohort, 99.8% of samples had a gestational age of less than 22 weeks (Supplemental Figure 4), which demonstrated no statistical relation to FF during this time period (Fig. 3a and Supplemental Figure 5A). This could differ from previous reports in that we only evaluated this association within the first trimester, which reportedly has a less rapid increase compared to later trimesters [5, 7, 9]. When following a small cohort of mothers, there have also been reports of a stable FF or even a temporary decreases in FF due to maternal weight gain during this gestational time [5]. Though FF may increase over the total pregnancy, these results demonstrate it is inconclusive if FF increases within statistical significance during the first trimester.
Fig. 3.
Male fetuses FFs as determined by DEFRAG_W as a function of gestational time (a), maternal age (b), weight (c), and trisomies (d)
We confirm previous reports that there is no correlation between FF and maternal age (Fig. 3b and Supplemental Figure 5B), but a negative correlation between FF and weight or BMI (Fig. 3c, Supplemental Figure 5c, and Supplemental Figure 6) [6–12]. It has been previously reported that trisomies 13 and 18 result in a decrease in FF, though there is some disagreement as to whether trisomy 21 fetuses result in a higher FF or not [7, 10–13]. While we observed a slight trend for increased FF in male trisomy 21 samples and a decrease in FF for trisomy 13 and 18 samples compared to euploid samples, this did not reach statistical significance (Fig. 3d).
Future larger cohorts will more effectively determine associations with FF. Specifically, this study was limited in the number of trisomy samples (56 trisomy 21, 10 trisomy 18, and 11 trisomy 13) and later gestational ages (32 samples > 21 weeks).
Summarizing previous results with these findings, we conclude that sample shipping time, maternal age, and gestational age (up to 21 weeks) has no effect on FF, whereas weight and BMI are negatively correlated with FF. Bioinformatics tools can have a profound impact on FF determination. We therefore propose developing future pipelines based on first running DEFRAG_W for determination of fetal sex and FF if male, followed by SeqFF for FF if the fetus is determined to be female. As methods vary by laboratory, individual laboratories should optimize their FF detection methods and not rely on arbitrary FF cutoff values for establishing reliable quality control metrics for the detection of fetal aneuploidies and smaller copy number variants with NIPS.
Electronic supplementary material
Acknowledgements
We are grateful to the VUmc NIPT team, the Dutch NIPT Consortium, and to all women who consented to participate in this study.
Compliance with ethical standards
Conflict of interest:
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0271-7) contains supplementary material, which is available to authorized users.
References
- 1.Takoudes T, Hamar B. Performance of non-invasive prenatal testing when fetal cell-free DNA is absent. Ultrasound Obstet Gynecol. 2015;45:112. doi: 10.1002/uog.14715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q, Cai Y, Brady P, Vermeesch JR. Real-time PCR evaluation of cell-free DNA subjected to various storage and shipping conditions. Genet Mol Res. 2015;14:12797–804. doi: 10.4238/2015.October.19.23. [DOI] [PubMed] [Google Scholar]
- 3.Peng XL, Jiang P. Bioinformatics approaches for fetal DNA fraction estimation in noninvasive prenatal testing. Int J Mol Sci. 2017;18:E453. doi: 10.3390/ijms18020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Beek DM, Straver R, Weiss MM, et al. Comparing methods for fetal fraction determination and quality control of NIPT samples. Prenat Diagn. 2017;37:769–73. doi: 10.1002/pd.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y, Zhou X, Huang S, et al. Quantitation of fetal DNA fraction in maternal plasma using circulating single molecule amplification and re-sequencing technology (cSMART) Clin Chim Acta. 2016;456:151–6. doi: 10.1016/j.cca.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Hudecova I, Sahota D, Heung MM, et al. Maternal plasma fetal DNA fractions in pregnancies with low and high risks for fetal chromosomal aneuploidies. PLoS ONE. 2014;9:e88484. doi: 10.1371/journal.pone.0088484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Zhu Z, Gao Y, et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod Sci. 2015;22:1429–35. doi: 10.1177/1933719115584445. [DOI] [PubMed] [Google Scholar]
- 8.Lee TJ, Rolnik DL, Menezes MA, McLennan AC, da Silva Costa F. Cell-free fetal DNA testing in singleton IVF conceptions. Hum Reprod. 2018;33:572–8. doi: 10.1093/humrep/dey033. [DOI] [PubMed] [Google Scholar]
- 9.Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33:662–6. doi: 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- 10.Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks’ gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41:26–32. doi: 10.1002/uog.12331. [DOI] [PubMed] [Google Scholar]
- 11.Suzumori N, Ebara T, Yamada T, et al. Fetal cell-free DNA fraction in maternal plasma is affected by fetal trisomy. J Hum Genet. 2016;61:647–52. doi: 10.1038/jhg.2016.25. [DOI] [PubMed] [Google Scholar]
- 12.Revello R, Sarno L, Ispas A, Akolekar R, Nicolaides KH. Screening for trisomies by cell-free DNA testing of maternal blood: consequences of a failed result. Ultrasound Obstet Gynecol. 2016;47:698–704. doi: 10.1002/uog.15851. [DOI] [PubMed] [Google Scholar]
- 13.Eiben B, Krapp M, Borth H, et al. Single nucleotide polymorphism-based analysis of cell-free fetal DNA in 3000 cases from Germany and Austria. Ultrasound Int Open. 2015;1:E8–E11. doi: 10.1055/s-0035-1555765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Schendel RV, Page-Christiaens GC, Beulen L, et al. Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part II-women’s perspectives. Prenat Diagn. 2016;36:1091–8. doi: 10.1002/pd.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oepkes D, Page-Christiaens GC, Bax CJ, et al. Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part I-clinical impact. Prenat Diagn. 2016;36:1083–90. doi: 10.1002/pd.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straver R, Sistermans EA, Holstege H, Visser A, Oudejans CB, Reinders MJ. WISECONDOR: detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42:e31. doi: 10.1093/nar/gkt992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straver R, Oudejans CB, Sistermans EA, Reinders MJ. Calculating the fetal fraction for noninvasive prenatal testing based on genome-wide nucleosome profiles. Prenat Diagn. 2016;36:614–21. doi: 10.1002/pd.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SK, Hannum G, Geis J, et al. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat Diagn. 2015;35:810–5. doi: 10.1002/pd.4615. [DOI] [PubMed] [Google Scholar]
- 19.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izenman A. Reduced-rank regression for the multivariate linear model. J Multivar Anal. 1975;5:248–64. doi: 10.1016/0047-259X(75)90042-1. [DOI] [Google Scholar]
- 21.Chan KC, Zhang J, Hui AB, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.