FIGURE 1.
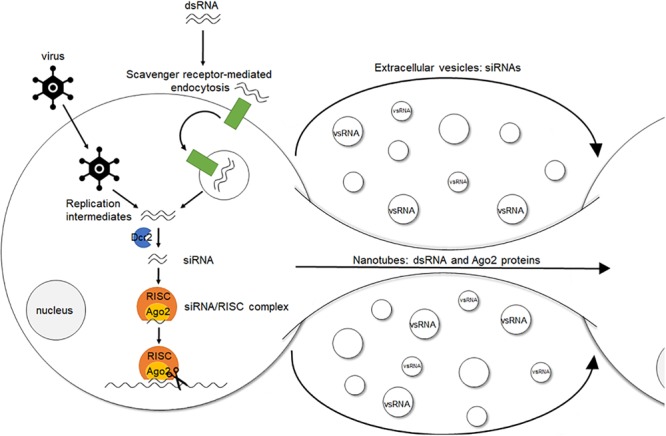
Simplified model of environmental, cell autonomous and systemic (antiviral) RNAi in insects. The siRNA pathway is triggered by dsRNA molecules. These duplexes naturally occur inside the cell during viral replication or can be artificially delivered. In the latter, cellular uptake of dsRNA, i.e., environmental RNAi, occurs via scavenger receptor-mediated endocytosis. Once inside the cell, i.e., cell autonomous RNAi, the dsRNA molecules are recognized in the cytoplasm and processed into siRNAs by Dcr2. Cleavage of viral RNA targets or endogenous transcripts is then further exerted by an Ago2-containing RISC, which encompasses the siRNA guide strand. Regarding antiviral RNAi in Drosophila, viral infection increases the formation of nanotube-like structures, through which short-distance transport of dsRNA and RISC components can occur. In addition, hemocyte-derived exosome-like vesicles systemically spread an antiviral RNAi signal (vsRNAs) in the hemolymph. This spread of the RNAi signal to cells in which the RNAi response had not been initiated before is named systemic RNAi. dsRNA, double stranded RNA. Dcr2, Dicer2. siRNA, small interfering RNA. RISC, RNA-induced silencing complex. Ago2, Argonaute2. vsRNA, viral small RNAs.
