Abstract
SETD5, a gene linked to intellectual disability (ID) and autism spectrum disorder (ASD), is a member of the SET-domain family and encodes a putative histone methyltransferase (HMT). To date, the mechanism by which SETD5 haploinsufficiency causes ASD/ID remains an unanswered question. Setd5 is the highly conserved mouse homolog, and although the Setd5 null mouse is embryonic lethal, the heterozygote is viable. Morphological tracing and multielectrode array was used on cultured cortical neurons. MRI was conducted of adult mouse brains and immunohistochemistry of juvenile mouse brains. RNA-Seq was used to investigate gene expression in the developing cortex. Behavioral assays were conducted on adult mice. Setd5+/− cortical neurons displayed significantly reduced synaptic density and neuritic outgrowth in vitro, with corresponding decreases in network activity and synchrony by electrophysiology. A specific subpopulation of fetal Setd5+/− cortical neurons showed altered gene expression of neurodevelopment-related genes. Setd5+/− animals manifested several autism-like behaviors, including hyperactivity, cognitive deficit, and altered social interactions. Anatomical differences were observed in Setd5+/− adult brains, accompanied by a deficit of deep-layer cortical neurons in the developing brain. Our data converge on a picture of abnormal neurodevelopment driven by Setd5 haploinsufficiency, consistent with a highly penetrant risk factor.
Introduction
The SET domain-containing family of HMTs encompasses the largest group of writers to the histone methylation code and is characterized by an evolutionarily conserved 130 amino acid SET domain. Human SETD5, located on chromosome 3p, stretches 23 exons in its full transcript and encodes the SET domain-containing protein 5, a 1442 amino acid protein. Its primary structure is highly conserved across vertebrates. Its role in development in general, and CNS development specifically, is increasingly appreciated. In 2012, the Jenuwein group purified SETD5 from HeLa extracts from a protocol to isolate HMTs, and recombinant mouse Setd5 was found to have H3K9 HMT activity1. Osipovich et al. found that mouse Setd5 co-immunoprecipitated with the NCoR complex of transcriptional regulators, and that Setd5−/− homozygous knockout animals were embryonic lethal beyond E11, due to multiple developmental defects including failure of neural tube closure and cardiac abnormalities2. Not only does SETD5 exert influence over critical elements of early development, but also it has been implicated in human neurodevelopmental disorders.
Converging evidence suggests that loss-of-function (LoF) mutations in SETD5 cause neurodevelopmental defects in humans3. SETD5 emerged as a candidate from bioinformatics analysis of a cohort of 996 patients in the United Kingdom with ID. Researchers found overlapping phenotypes in ID patients harboring heterozygous SETD5 LoF mutations and the existing 3p microdeletion syndrome, which spans SETD5 on the short arm of chromosome 34. A subsequent study performing whole exome sequencing (WES) from a cohort of 250 ID patients found seven patients with missense, nonsense, splice-site, or microdeletion SETD5 mutations5. This study attributed the microdeletion patients’ phenotype to nonsense-mediated decay (NMD) based on reduced SETD5 mRNA transcript levels in HEK293 cells transfected with CRISPR/Cas9 guide-RNAs (gRNA) recapitulating the patients’ microdeletion5. Pinto et al. identified SETD5 as a candidate autism spectrum disorders (ASD) susceptibility gene based on the discovery of de novo copy number variants (CNVs) observed in a cohort of 2446 ASD patient genomes6.
We took advantage of the Setd5+/− heterozygous mouse as a model of Setd5 LoF. We hypothesized that LoF would be particularly pronounced in neurons from cerebral cortex, which is widely implicated in ASD pathogenesis7 and a cell type enriched in SETD5/Setd5 expression8. To elucidate the role of Setd5 in nervous system development and its contributions to ASD/ID, we studied the morphological and electrophysiological features of dissociated cortical neurons, the molecular and transcriptomic signature underlying Setd5 reduction, neuroanatomical details of adult and developing brains, and finally, behavioral phenotypes of adult animals.
Materials and Methods
Primary cortical neuron culture
Primary cortical neurons were harvested from E18.5 or P0 animals as described previously9 and grown in vitro on glass. Ara-C was used to arrest proliferation of glial progenitors in neuronal cultures.
Immunohistochemistry/immunocytochemistry
Mouse brain tissue for immunohistochemistry was collected by perfusion-fixation and frozen sections. Sections were permeabilized, blocked, and incubated in primary antibody solution overnight (Table 1), followed by a fluorescently-labeled secondary antibody and nuclei staining. For cells, they were fixed, permeabilized, blocked then incubated in primary antibody solutions overnight, followed by a fluorescently-labeled secondary antibody and nuclei staining. Neuron morphology was assessed by Neurolucida tracing. Synapses were quantified by the co-localization of pre- and post-synaptic puncta as previously described10. Cell death was quantified by TUNEL staining. For all assays, unstained controls were used to ensure no background GFP staining was observed in Setd5+/− samples.
Table 1.
List of all antibodies used in the study
| Application | Antibody | Dilution | Manufacturer | Catalog |
|---|---|---|---|---|
| Immunocytochemistry | MAP2 | 1:2000 | Abcam | 5392 |
| CTIP2 | 1:500 | Abcam | 25B6 | |
| VGLUT1 | 1:500 | Synaptic systems | 135011 | |
| HOMER1 | 1:500 | Synaptic systems | 160003 | |
| GFAP | 1:1000 | Dako | Z0334 | |
| NEUN | 1:1000 | Millipore | 377 | |
| GAD65/67 | 1:1000 | Abcam | 11070 | |
| GFP | 1:1000 | Abcam | 290 | |
| Immunohistochemistry | BRN2 | 1:500 | Santa Cruz | 6029 |
| TBR1 | 1:500 | Abcam | 31940 | |
| CTIP2 | 1:500 | Abcam | 25B6 | |
| PSD95 | 1:500 | Abcam | 18258 | |
| SATB2 | 1:500 | Abcam | 51502 | |
| Flow cytometry | CD24-PE | 1:3000 | Abcam | 218742 |
| CD45-AF647 | 1:600 | Biolegend | 103124 |
Electrophysiology
Primary cortical neurons were cultured on Axion Maestro multielectrode array (MEA) plates and spontaneous spike activity recorded serially starting at DIV. LFP computation was obtained by a custom MATLAB algorithm to calculate the power ratio at 1–10 and 100–150 Hz ranges. For imaging of calcium transient activity, cells were loaded with the calcium-sensing dye Fluo-4AM and spontaneous firing activity was recorded.
Electroencephalogram
First, spontaneous EEG activity was recorded in awake behaving adult mice via hippocampal-implanted electrodes. Next, seizure threshold was assessed by measuring latency to epileptic spikes provoked by the convulsant PTZ.
RNA-Seq/scRNA-Seq
Primary cortical neurons from E18.5 fetuses were extracted as described11. Once in single-cell suspension, they were stained with pan-neuronal marker CD24 and hematopoietic marker CD45 and sorted by fluorescence-activated cell sorting (FACS). Live neuronal cells were collected and RNA libraries prepared and sent for bulk RNA-Seq. Libraries were sequenced and gene ontology (GO) analysis conducted of differentially expressed genes. For single-cell (sc) RNA-Seq, the same sorting protocol was used, but cells were then used to create single-cell libraries prior to sequencing. t-SNE plots were generated and cells clustered. The most highly expressed genes and differentially expressed genes within the clusters were analyzed.
Animals
The Setd5GFP transgenic mouse2, hereafter referred to as the Setd5+/− heterozygous knockout animal was used for all animal experiments in accordance with the Institutional Animal Care & Use Committee (IACUC) at UCSD and Cleveland Clinic.
Neurologic and metabolic assays
Weight, grip strength, rotorod12, and composite neurological score13 were measured at age 10 weeks.
Behavioral assays
Behavioral experiments were conducted 1 week apart to prevent test fatigue, using two independent cohorts. The open field test was conducted as described previously12. The Barnes maze was conducted as described previously14. The elevated plus maze was conducted as described previously15. Overnight nest building was performed as described previously14.
3-chamber social interaction
The 3-chamber test was conducted as described previously14,16 in three subtrials: sociability (novel animal vs. empty), social novelty (novel animal vs. familiar), and social preference (cagemate animal vs. novel).
Magnetic resonance imaging
Brain MRI was performed of fixed adult mouse skulls perfused with gadolinium-containing contrast aent as previously described17. The volume of various brain regions was assessed and compared by genotype.
Microscopy
Fluorescent microscope images were acquired on a Zeiss Apotome microscope using Zen software.
Statistics
Detailed statistical methods are included in the description of individual experiments. Statistical analysis was conducted with GraphPad Prism 7 software, generally by t-test or 2-way ANOVA with α = 0.05. A false discovery rate (FDR) significance value of 0.05 was used to for tests with multiple comparisons.
Results
Morphological alterations and hypoconnectivity in Setd5+/− primary cortical neurons
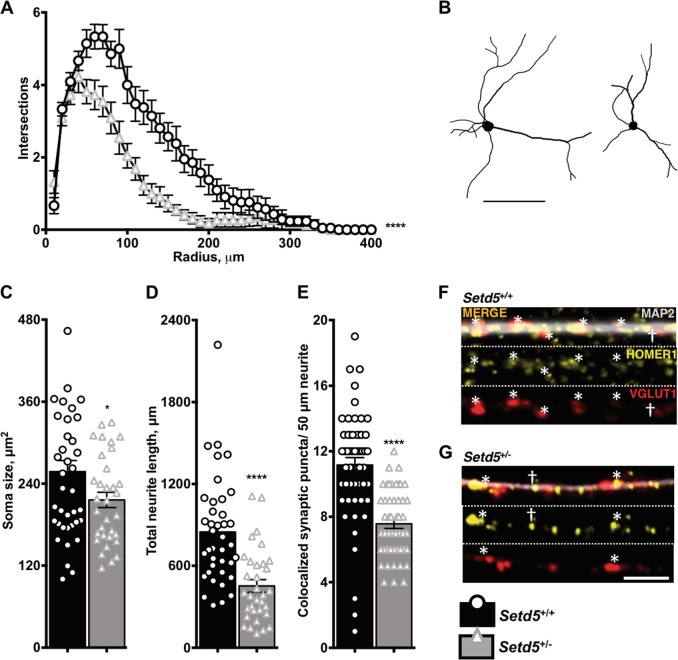
Alterations in neuronal morphology and function were previously described for other ASD-related chromatin regulator genes9. Thus, we cultured primary cortical neurons harvested from P0 Setd5+/+ and Setd5+/− animals in vitro for 24 days. Ctip2+ cells were selected for analysis as the predominant projection neurons in the cultures with extensive measurable arborization. WT (left) and het (right) neurons are depicted (Fig. 1b, scale bar 100 μm); when subjected to Sholl analysis, het neurons were found to have significantly fewer intersections with 10 μm concentric circles (F1,1640 = 169.8, P < 10−4, Fig. 1a). Setd5+/− neurons also had significantly smaller soma size (257.7 ± 16.2 vs. 216.2 ± 11.19 μm2, P = 0.0461, Fig. 1c) and significantly reduced neuritic outgrowth (848.1 ± 62.41 vs. 452.2 ± 46.27 μm, P < 10−4, Fig. 1d) despite no significant differences in total neurite number (3.282 ± 0.127 vs. 3.091 ± 0.1649, P = 0.3545, data not shown). When stained by immunocytochemistry for the pre- and post-synaptic markers VGLUT1 and HOMER1, respectively, Setd5+/− neurons had reduced density of colocalized pre- and post-synaptic puncta on MAP2+ neurites (11.16 ± 0.4508 vs. 7.569 ± 0.2818 puncta/50 μm, P < 10−4, Fig. 1e–g, scale bar 5 μm). To determine whether differences in cell viability contributed to the observed morphological differences, fixed cells were subject to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL). No difference in percent apoptotic neurons was detected (14.57 ± 2.895 vs. 10.21 ± 2.254 percent, P = 0.3220, Fig. S1A). To determine whether Setd5 LoF impacted excitatory:inhibitory balance, a purported aspect of aberrant network connectivity in ASD18, fixed cultures were stained for the excitatory and inhibitory markers VGLUT1 and GAD65/67, respectively. No differences in percent VGLUT1+ cells (77.09 ± 4.389 vs. 70.87 ± 3.457 percent, P = 0.2821, Fig. S1B) or percent GAD65/67+ cells (32.5 ± 4.22 vs. 38.52 ± 3.154 percent, P = 0.2700, Fig. S1B) were detected. To exclude any effects of culture heterogeneity on morphology and synaptogenesis observed in neurons, cell cultures were fixed and stained for the mature postmitotic neuron marker NeuN and glial marker GFAP, in which no differences in percent neurons (82.55 ± 3.10 vs. 80.72 ± 4.04 percent, P = 0.7181, Fig. S1C) or glia (17.45 ± 3.10 vs. 19.28 ± 4.04 percent, P = 0.7181, Fig. S1C) were observed. As a further inquiry into culture heterogeneity, no genotype-dependent difference in percent positive CTIP2+ neurons was detected (48.43 ± 3.40 vs. 51.60 ± 4.11 percent, P = 0.5571, Fig. S1D).
Fig. 1. Setd5+/− primary cortical neurons display signs of morphological alterations consistent with hypoconnectivity.
a Het neurons present significantly fewer intersections by Sholl analysis (genotype ****P < 10−4, F1,1640 = 169.8, DF = 1). b Representative traces of wt (left) and het (right) neurons are depicted (scale bar 100 μm). c Het neurons have significantly smaller soma (*P = 0.0461, t1,70 = 2.031, DF = 70). d Total neuritic outgrowth is significantly reduced among het neurons (****P < 10−4, t1,70 = 4.94, DF = 70). e HOMER1+ VGLUT1+ colocalized synaptic puncta are significantly less dense in MAP2+ neurites of het neurons (****P < 10−4 t1,104 = 6.643, DF = 104). f–g Representative image of wt and het neurite synaptic puncta quantification; top = merge MAP2 (white), VGLUT1 (red), HOMER1 (yellow); center = VGLUT1 (red); bottom = HOMER1 (yellow); *asterisks represent colocalized synaptic puncta, while †dagger represents uncounted non- colocalized punctum, scale bar 5 μm. Statistics: n = neurons from 12 total animals per genotype from three total experiments. (a–d) = 39 Setd5+/+ and 33 Setd5+/− CTIP2+ neurons, with group means compared by t-test (b–d) or 2-way ANOVA (a) for factors radius and genotype. (f) = 55 Setd5+/+ and 51 Setd5+/− neurons, with group means compared by t-test. Replicates as individual neurons (a–e) with error bars representing mean ± SEM
Delayed network development in Setd5+/− cortical neurons
To ascertain the functional consequences of altered morphology and hypoconnectivity among Setd5+/− cortical neurons and investigate a potential cellular basis for epileptogenesis in SETD5 human patients, cells were cultured atop a multielectrode array (MEA), with serial recordings obtained beginning at 8 days in vitro (DIV, Table 2). Representative raster plots of spontaneous spike activity from 21 DIV are depicted (Fig. 2a, scale bar 20 s). When spontaneous neural recordings were analyzed, Setd5+/− neurons had significantly reduced weighted mean firing rate (P < 10−4, Fig. 2b). Bursting activity also had genotype-dependent differences: Setd5+/− neurons had significantly reduced normalized burst frequency (P = 0.0002, data not shown), with bursts spreading to significantly fewer electrodes (P < 10−4, data not shown). Moreover, Setd5+/− neuron firing was significantly less synchronous (P = 0.0004, Fig. 2c). To ensure spiking originated from neuronal-initiated action potential firing, 10 nM TTX was added to the cultures, which completely abolished spiking (data not shown). Complementary to the spiking results, local field potential (LFP) recordings obtained from raw MEA data (Fig. 2d) showed genotype-dependent low- and high-frequency power changes (Fig. 2g), providing evidence for network-level differences in activity. Low-frequency (1–10 Hz) transients in the LFP—markers of network synchronized depolarization19—were not significantly different between genotypes overall, although intervals of difference did emerge, especially within the second week in culture (6–11 DIV, Fig. 2e). Interestingly, high-frequency (100–150 Hz) power—sometimes known as broadband or high gamma power, an index of local circuit firing rate mostly provided by neuronal activity20—was significantly different between genotypes (P = 0.0042, Fig. 2f), with particularly pronounced differences in the 6–11 DIV critical period.
Table 2.
Recording parameters for multielectrode array
| Axion maestro multielectrode array recording protocol | |
|---|---|
| Configuration | Real-time spontaneous neural |
| Length | 30 min |
| Spike activity criteria | Field potential ≥ 6σ above noise |
| Active electrode criteria | 5 spikes•min−1 |
| Burst criteria | Spikes in ≥ 5 active electrodes |
Fig. 2. Setd5+/− cortical neurons trail wt counterparts in development of synchronized neural networks.
a Raster plots depict spontaneous firing activity in eight electrodes from a single well (wt, top; het, bottom) showing a qualitatively different firing pattern (scale bar 20 s). b Spontaneous spike activity is significantly reduced in het cells (genotype ****P < 10−4, F1,341 = 22.37, DF = 1). c Synchrony index among het neurons lags significantly behind wt (genotype ***P < 10−3, F1,379 = 12.86, DF = 1). d Example LFP trace after low-pass filtering raw MEA signal at 500 Hz. e, f Log10 power ratio baselined to first day of MEA recording for low-frequency (1–10 Hz, e) and high-frequency/broadband gamma (100–150 Hz, f) power, with genotype-dependent differences in power ratio for high-frequency only [(entire time period: low-frequency F1,45 = 0.55, P = 0.4644 and high-frequency F1,45 = 10.17, **P = 0.0042); (6–11 DIV only: low-frequency F1,11 = 18.91, **P = 0.0074 and high-frequency F1,11 = 33.36, **P = 0.0022)]. g Example power spectral densities (PSD) from Setd5+/+ (black) and Setd5+/− (gray) cultures. Blue and yellow boxes denote low- and high-frequency regions of interest for (e) and (f), respectively. h Significantly reduced correlated firing activity among het neurons (****P < 10−4, t1,12 = 10.65, DF = 8) measured by Ca2+ transients. Statistics: (b, c) n = neurons from 12 Setd5+/+ and Setd5+/− animals each; 2-way ANOVAs for factors genotype and DIV; replicates = individual wells (neurons from 1 animal/well) pooled from three separate experiments. (e, f) Replicates defined as data from neurons of individual animals, n = 8 Setd5+/+ and n = 4 Setd5+/−; 2-way ANOVAs for factors genotype and DIV. h n = neurons from five animals per genotype (583 Setd5+/+ or 505 Setd5+/− active total neurons), with each animal as a single replicate and t-test of group means. Error bars representing mean ± SEM
Quality controls to ensure culture heterogeneity did not contribute to network-level electrophysiological differences were conducted. While cell viability was shown to be comparable between genotypes (Fig. S1A), to ensure that variation in cell density did not contribute to network connectivity, cells were imaged and observed at comparable coverage of the MEA (Fig. S1E, F), and total protein collected from each well was comparable between genotypes (P = 0.6142, data not shown).
While MEA analysis provides a global overview of network connectivity, it lacks single-cell optical resolution afforded by other modalities. Calcium signaling in neurons mediates several critical functions including synaptic plasticity, and calcium channelopathies have been implicated in select monogenic neurodevelopmental conditions such as Timothy syndrome21,22. To determine whether Setd5 haploinsufficiency impacted calcium signaling in cortical neurons, we employed optical recordings using a fluorescent dye to monitor calcium transients (Fig. S2A, B, scale bar 50 μm) and quantified the neuronal traces (three representative traces, Fig. S2C). Neurons from Setd5+/− cultures demonstrated reduced synchrony of spontaneous transients, with significantly fewer neurons participating in correlated firing activity (0.292 ± 0.0454 vs. 0.053 ± 0.021 correlated firing response ratio, P < 10−4, Fig. 2h).
Developing brain single-cell transcriptomic analysis
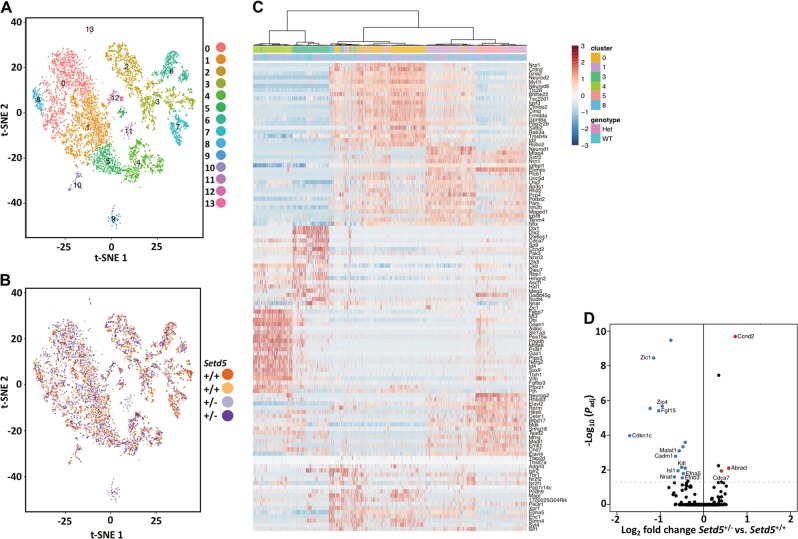
We next focused on potential molecular differences in the neurons from the mouse brain. We performed bulk RNA-Seq of sorted neuronal cells (CD24+, CD45−, Fig. S3) from the developing (E18.5) cortex. Gene ontology analysis of differentially expressed (DE) genes (82 total with FDR P < 0.05) revealed 20 ontological categories (Fig. S4). Although extracellular matrix-related genes were significantly downregulated in Setd5+/− cells, this did not fully reveal a transcriptomic basis for the morphological and electrophysiological differences we observed in Setd5+/− neurons (Supplemental Sheet 1). We repeated the dissection and sorting, then used single-cell (sc) RNA-Seq to reveal potential genotype-dependent differences in the developing cortex cellular composition. Pooled data from both genotypes was clustered using the Seurat R package23, and the resulting 14 clusters were displayed using t-distributed stochastic neighbor embedding (t-SNE, Fig. 3a, b). The most highly expressed genes in each cluster are listed (Supplemental sheet 2). Clusters 0, 1, 3, 4, 5, and 8 were identified for more extensive heatmap analysis (Fig. 3c) based on enrichment of neurodevelopmental genes among the top 20 expressed genes. Hierarchical clustering separated these six clusters (Fig. 3b). Cluster 3 had the greatest number of significantly (FDR P < 0.05) DE genes by genotype. Literature search of Cluster 3 DE genes revealed effects of Setd5 haploinsufficiency in upregulation (Ccnd2, P < 10−9; Abracl, P = 0.0080; Nr2f1, P = 0.0155; Cdca7, P = 0.0503) or downregulation (Zic1, P < 10−8; Zic4, P < 10−5; Fgf15, P < 10−5; Malat1, P = 0.0008; Cadm1, P = 0.0016; Pnoc, P = 0.0080; Kitl, P = 0.0084; Efna5, P = 0.0161; Nnat, P = 0.0318; P = 0.0290, Fig. 3d, Supplemental Sheet 1) of select genes related to nervous system development. Relative expression by cluster and genotype of select neural marker genes is also depicted (heatmap, Fig. S5A, split-dot plot, Fig. S5B).
Fig. 3. Single-cell transcriptomic profiling of E18.5 Setd5+/− cortical progenitors.
a, b Whale-octopus plots of t-distributed stochastic neighbor embedding (t-SNE) dimensional reduction of barcoded single-cell RNA-Seq reads into 14 clusters (a) distributed across both genotypes (b). c Hierarchical clustering of Z-scaled single-cell expression values reveals separation of t-SNE clusters and distribution of genotypes within individual clusters. d Volcano plot of cluster 3 genes showing differentially expressed genes in Setd5+/− (significantly downregulated: blue; significantly upregulated: red; select genes of interest labeled) Statistics: Sorted CD24+ CD45− cortical suspensions from n = 2 E18.5 fetuses per genotype. n = 2066–2858 cells, 39805–54534 reads/cell (Setd5+/+) and 2269–2645 cells, 48174–56267 reads/cell (Setd5+/−)
Setd5+/− animals have abnormal patterns of social interaction and demonstrate autism-compatible behaviors
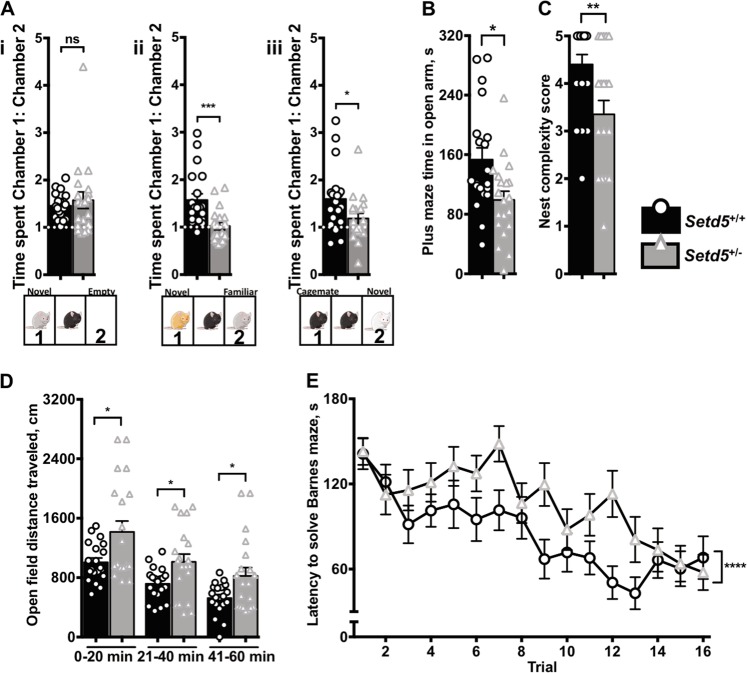
We subjected independent cohorts of adult animals to established behavioral assays in animal models of ASD15,24. The 3-chamber social interaction test captures differences in the experimental animal’s preference for interacting with a known versus novel animal and is frequently employed in animal models of autism. Although Setd5+/− animals had no defects in social approach compared with controls (1.457 ± 0.060 vs. 1.575 ± 0.175 ratio of time with novel mouse vs. empty chamber, P = 0.5274, t1,38 = 0.6379, Fig. 4ai), they lacked the normal preference for novel over familiar (1.573 ± 0.130 vs. 1.021 ± 0.078, ratio of time with novel mouse vs. familiar mouse, P = 0.008, t1,38 = 3.643, Fig. 4aii) in the social novelty preference test. This trend was repeated in the test of social preference for cagemate versus novel mouse (1.599 ± 0.149 vs. 1.187 ± 0.108, ratio of time time with cagemate mouse vs. novel mouse, P = 0.031, t1,38 = 2.24, Fig. 4aiii), suggesting that het animals failed to distinguish known from unknown conspecifics. Setd5+/− mice spent significantly reduced time in the open arms of the elevated plus maze versus wt controls (153.3 ± 15.91 vs. 99.25 ± 11.81 s, P = 0.0096, Fig. 4b), suggestive of increased anxiety. They also built significantly less complex nests in the overnight nest-building task (nest scores 4.4 ± 0.2103 vs. 3.35 ± 0.2927, P = 0.0074, Fig. 4c). We also conducted an open field test, finding significantly elevated activity levels among Setd5+/− animals over the course of the 1 h test (t-test P values 0.0135 0–20 min; 0.0148 21–40 min; 0.0175 41–60 min; ANOVA genotype P < 10−4, Fig. 4d). Thigmotaxis, the tendency of rodents to prefer the perimeter of the open field task, was also affected: genotype was found to be a significant factor accounting for variation in time spent in the central 25% of the chamber (genotype *P = 0.0343, Table 3). Finally, the Barnes maze task of cognition and spatial memory and recall was conducted: Setd5+/− animals displayed significantly reduced performance over the course of the 16 trials versus wt controls (P < 10−4, Fig. 4e). Complete data when separated by sex (Fig. S6) and analyzed by ANOVA (Table 3) are displayed. Gross neurological deficits in the het animals were ruled out by the observation of comparable performance on rotorod, grip strength, and composite neurological tests13,25 (Fig. S7).
Fig. 4. Setd5+/− animals display autism-compatible behaviors. Pooled male-female data of behavioral tasks illustrating genotype-dependent differences.
a (i) 3-chamber social interaction test. Left: No genotype-dependent differences in social approach as measured by ratio of time spent in chamber with novel animal vs. empty chamber (1.457 ± 0.060 vs. 1.575 ± 0.175, P = 0.5274, t1,38 = 0.6379). (ii) Center: Het mice lack normal preference for novel over familiar animal as measured by ratio of time spent in chamber with novel animal vs. familiar animal (1.573 ± 0.130 vs. 1.021 ± 0.078, **P = 0.008, t1,38 = 3.643). (iii) Het mice lack normal preference for cagemate over novel animal as measured by ratio of time spent in chamber with cagemate animal vs. novel animal (1.599 ± 0.149 vs. 1.187 ± 0.108, *P = 0.031, t1,38 = 2.24). b Het mice spend significantly less time in open arms of elevated plus maze (*P = 0.0112, F1,36 = 7.146, DF = 1). c Het mice build significantly less complex nests in overnight nest-building task (genotype **P = 0.0074, Mann–Whitney ranked U = 105.5, DF = 1). d Het mice are hyperactive in open field (genotype ****P < 10−4, F1,2 = 18.27, DF = 1). e Het mice are impaired on Barnes maze (genotype ****P < 10−4, F1,15 = 21.8, DF = 21.8). Statistics: n = 10 total animals per sex, per genotype; t-test of pooled male-female data (a, b, d); Mann–Whitney ranked test (c); 2-way ANOVA (e). Graphs are representative of pooled male and female data for each genotype, with replicates as individual animals and error bars mean ± SEM
Table 3.
Complete statistics for behavioral experiments
| Behavioral task | Genotype P | Genotype F | Sex P | Sex F |
|---|---|---|---|---|
| Sociability | 0.5317 | 0.3989 | 0.2909 | 1.149 |
| Social-novelty preference | 0.004 | 15.06 | 0.0131 | 6.821 |
| Social-preference | 0.0299 | 5.1112 | 0.384 | 0.7768 |
| Elevated plus maze | 0.0112 | 7.146 | 0.91 | 0.013 |
| Nest building | 0.0071 | 8.150 | 0.8296 | 0.0185 |
| Barnes maze | <0.0001 | 21.80 | 0.0002 | 13.65 |
| Open field | <0.0001 | 18.27 | 0.3495 | 0.8829 |
| Open field time in center | 0.0343 | 4.891 | 0.0103 | 7.433 |
Epileptic activity is a known feature of certain SETD5 LoF patients3. Thus we subjected adult mice to electroencephalogram (EEG) via hippocampal-implanted electrodes to determine whether the animal model recapitulated patient seizures. Neither Setd5+/+ nor Setd5+/− animals showed spontaneous seizure activity (Fig. S8A, B), and no genotype-dependent differences in seizure threshold were detected when sub-convulsive doses of PTZ (pentylenetetrazol) were successively injected prior to recording (P = 0.6549, Fig. S8C–F).
Anatomical changes in adult mouse brain by MRI
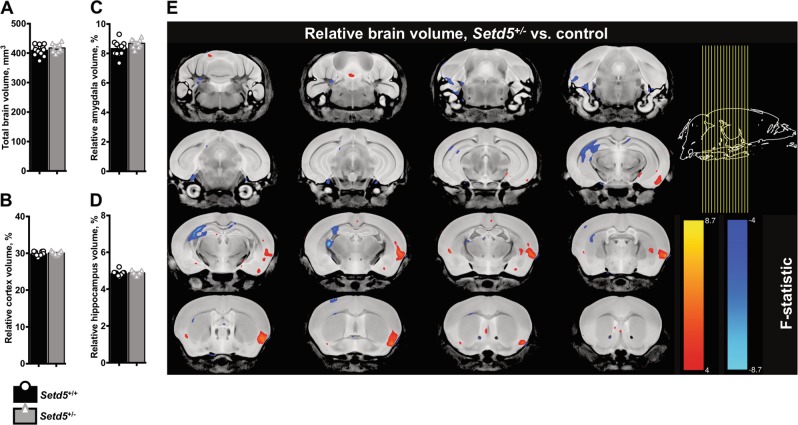
Region-specific differences in brain volume and connectivity are frequently observed in human ASD patients and animal models by magnetic resonance imaging (MRI)26,27. Given that Setd5+/− animals recapitulated select behaviors compatible with the human phenotype, anatomical imaging was undertaken to ascertain neuroanatomical-neurobehavioral correlations. Absolute and relative differences in brain volume were calculated by analysis of MRI from adult mouse brains. No difference beyond the FDR P < 0.05 was observed in total brain volume (P = 0.2606, Fig. 5a), total relative cortical volume (P = 0.1910, Fig. 5b), relative amygdala volume (P = 0.0471, FDR P = 0.21, Fig. 5c), or total relative hippocampus volume (P = 0.4131, Fig. 5d). However, when individual subregions were normalized to total brain volume, significant genotype-dependent differences were observed in several specific regions (Fig. 5e). Brain regions whose relative size differences exceeded the false discovery rate (FDR) P = 0.05 are overlaid in the coronal series (blue = smaller, red = larger, Fig. 5e). For absolute size differences not normalized to total brain volume, see Fig. S9. Specifically, the following autism-relevant brain regions emerged as regions of significant difference in Setd5+/− brains beyond the FDR: secondary auditory cortex28 (larger, FDR P = 0.03), dorsolateral orbital cortex (larger, FDR P = 0.01) and frontal association cortex29 (larger, FDR P = 0.03). In the hippocampus, differences in the CA2 cell layer fell short of statistical significance (FDR P = 0.06). A comprehensive list of all subregions is displayed in Supplemental Sheet 3.
Fig. 5. Anatomical differences in brain region volume in Setd5+/− adults.
a No genotype-dependent difference in brain volume (409.7 ± 5.29 vs. 417.5 ± 4.16 mm3, t1,21 = 1.156, P = 0.2606). b No genotype-dependent difference in relative cortex volume (29.81 ± 0.16 vs. 30.09 ± 0.13 mm3, t1,21 = 1.35, P = 0.1910). c Genotype-dependent difference in relative amygdala volume does not exceed FDR (8.331 ± 0.145 vs. 8.693 ± 0.082 percent, t1,21 = 2.109, FDR P = 0.21). d No genotype-dependent difference in relative total hippocampus volume (4.842 ± 0.042 vs. 4.885 ± 0.0288 percent, t1,21 = 0.8351, P = 0.4131). e Coronal series highlighting regional relative size differences with significance exceeding FDR threshold = 0.05 (blue = smaller, red = larger). Statistics: n = 12 (wt) or n = 11 (het) adult brains as independent replicates, with bars representing mean ± SEM. Brain region size normalized to total brain volume. t-test for comparison of group means of 178 brain regions, with statistical significance assigned at F ≥ 4.0, corresponding to FDR P ≤ 0.05
Postnatal brain deficit in CTIP2+ cortical layer
We next performed a more detailed anatomical investigation. Evidence of aberrant cortical lamination has been observed in mouse models17,18 and proposed in human ASD7. To determine whether Setd5+/− behavioral abnormalities in adults could be attributed to early lamination defects, we chose P1 and P10 brain sections stained for superficial cortical neuron marker Brn2 (purple) and deep cortical neuron markers Ctip2 (green) and Tbr1 (red)30,31 as described previously17 (Fig. S10A, B) for the ability to track development of cellular subpopulation composition. Cell layers were measured in images, at least 2 per animal, of coronal sections corresponding to the Allen Developing Mouse Brain Reference Atlas sections 90–14032 containing M1, at 30 and 70% from the dorsal midline. In P1 brain sections, no differences in thickness of the deepest or most superficial layers were observed: (TBR1+ layer, P = 0.9090; BRN2+ layer, P = 0.5906, Fig. S10C, G), although the CTIP2+ layer was significantly thinner in Setd5+/− sections (144.2 ± 7.78 vs. 113.9 ± 6.25 μm, P = 0.0055, Fig. S10E). Similarly, in P10 brain sections, no differences in thickness of the deepest or most superficial layers were observed (TBR1+ layer, P = 0.0945, Fig. 6D; BRN2+ layer, P = 0.0883, Fig. S10G), although the CTIP2+ layer was again significantly thinner in Setd5+/− sections (214.3 ± 7.11 vs. 178.4 ± 6.99 μm, P = 0.0012, Fig. S10E). Despite the differences in total layer thickness, no genotype-dependent differences were detected in cell density within the respective cortical layers at P1 or P10 (TBR1+ layer, P = 0.7481, P = 0.2191 Fig. S10D; CTIP2+ layer, P = 0.1064, P = 0.4667, Fig. S10F; BRN2+ layer, P = 0.5256, P = 0.1166 Fig. S10H).
Discussion
SETD5 has emerged as an intriguing ASD-risk candidate, guiding us to investigate the Setd5 animal model to shed light on the contributions of this gene to neurodevelopment. Construct validity of the Setd5 mouse with human ASD patients harboring SETD5 mutations is of critical significance in extrapolating the findings of the animal model to human ASD etiology.
Findings from our in vitro and in vivo experiments echo published reports of existing animal models of autism and extend those findings at the level of physiology, which in the Setd5+/− neurons and brain appear to converge on a picture of cortical hypoconnectivity. Synaptogenesis is impacted in various animal models of autism, from significant decreases in excitatory post-synaptic currents (mESPCs) among CA1 hippocampal neurons in neurexin-1α mice33 to increased synaptic density among cortical neurons in Fragile-X mice34. Cortical neurons from Setd5+/− animals demonstrated reduced excitatory synaptic density. Importantly, this was not attributed to culture heterogeneity, as the CTIP2+ proportion of cells was comparable across genotypes in the current study. We observed significantly reduced neuritic outgrowth and soma size in cultured Setd5+/− deep-layer cortical projection neurons. These deficits affected cortical layer V neurons positive for CTIP2, a population of cortical subprojection neurons implicated in human ASD pathogenesis7, which were found as a point of convergence among distinct ASD-related mutations35.
These data were corroborated by the reduced synchrony of network firing in Setd5+/− cortical neurons. Significant genotype-dependent differences in high-frequency activity were observed over time, with focal differences in both low- and high-frequency activity specifically in the second week in vitro. These findings suggest a critical period of in vitro maturation during the second week in culture, in which control cultures showed both higher and more synchronized network activity, potentially leading to a more mature circuit via activity-dependent synapse formation and a sustained developmental difference persisting into the future.
Our transcriptomic analysis identified a subtle effect of Setd5 haploinsufficiency on a specific cellular subpopulation in the developing brain, as assessed by differential gene expression in Setd5+/− cortical neuronal cells. At the level of bulk RNA-Seq, downregulation of genes related to extracellular matrix organization was observed. A number of genes involving collagen synthesis, which is known to be dysregulated in models of ASD36, were also downregulated. Interestingly, the majority of these neurodevelopmental downregulated genes affected by Setd5+/− occurred in cluster 3, a population of cells characterized by expression of inhibitory interneuron markers Dlx1/Dlx237 and Cdca7, a gene that regulates intermediate progenitor production in the developing brain38. Significant downregulation of Nnat expression, involved in neuronal intracellular calcium signaling39, could connect to our finding of reduced calcium transient synchrony in Setd5+/− cells. Cdkn1c, encoding cell cycle regulator p57, mediates cortical neurogenesis40 and was also significantly downregulated in the Setd5+/− cells. Malat1, a long non-coding RNA (lncRNA) that was significantly downregulated in the Setd5+/− cells, contributes to neurite outgrowth through the MAPK pathway during neuronal differentiation41 and could connect to our findings of neurite outgrowth restriction in Setd5+/− neurons. Further analysis of Setd5 deletion in specific cell types will elucidate the significance of Setd5-mediated transcriptomic abnormalities in the developing brain.
Extensive anatomical imaging data are available for both human ASD patients and genetic animal models. The most recent mega-analysis of human ASD MRI data found consistently decreased volume of the putamen, amygdala, and nucleus accumbens; and increased frontal but decreased temporal cortex thickness42. Setd5+/− brains showed no evidence of overall macrocephaly, although significant volume differences were observed in several ASD-implicated brain subregions including regions of frontal cortex important for learning. An MRI analysis of 26 mouse models found consistent abnormalities of parieto-temporal and frontal lobes, cerebellar cortex, hypothalamus, and striatum, clustering the known models into three groups based on correlated size changes in distinct brain regions26. Setd5+/− brains lacked the hallmarks of Ellegood et al.’s Group 1 (increased large white matter tract size) and Group 3 (decreased frontal cortex size) and thus may cluster approximately in Group 2 (decreased hippocampal size)26. These data provide an interesting insight into the downstream neuroanatomical consequences of Setd5 haploinsufficiency, though it is premature to claim a causal relationship with observed behavioral phenotypes as the studies were not conducted in identical animals (i.e., correlating an individual animal’s behavior results with postmortem MRI analysis)17.
The Setd5+/− adult animal demonstrated features compatible with human ASD, including those manifest by SETD5 LoF human patients3,4. The three-chambered test of social interaction captures differences in social interaction preferences in numerous animal models of ASD24. Although Setd5 het animals had normal social approach tendency, when faced with the task of discriminating between a familiar versus unfamiliar conspecific, mutant animals showed no preference for novel versus familiar, in contrast to controls. This pattern differs from other animal ASD models in that reduced sociability and lack of preference for social novelty were frequently observed together, including in the Nlgn4, Slc6a4, and Gabrb3 models24, thus pointing to a more specific deficit in social recognition, rather than globally impaired socialization in the Setd5+/− animals. The Barnes maze task detected a mild cognitive and spatial learning deficit in Setd5+/− animals, a behavioral domain implicated in other ASD models43. Finally, the open field test revealed hyperactivity consistent with other mouse ASD models44, although the reduced thigmotaxis evidenced by increased time in the central area of the chamber, differed from other ASD models45,46.
In conclusion, we report here a new animal model for ASD, with autism-compatible CNS phenotypes at the cellular, network, and behavioral levels. Importantly, the animal model has validity with observed human ASD patients harboring SETD5 mutations. The present model suggests that Setd5 haploinsufficiency has a negative impact on early brain development, potentially through aberrant gene expression in select neuronal subpopulation(s) that contributes downstream to functionally hypoconnected cortical networks and behavioral abnormalities. Our data yield novel insights into the pathogenic basis for SETD5 LoF ASD patients and provide a model for investigating the contribution of ASD susceptibility genes.
Supplementary information
Acknowledgements
This work was supported by the National Institutes of Health through the R01MH108528, R01MH094753, R01MH109885, R01MH100175 and U19MH107367, part of the National Cooperative Reprogrammed Cell Research Groups (NCRCRG) to Study Mental Illness. We would like to acknowledge the Albert La Spada group at UCSD for their generous use of animal behavior equipment. We would like to acknowledge the Human Embryonic Stem Cell Core Facility at UCSD/Sanford Consortium for Regenerative Medicine for use of the MEA machine. S.M.M. was partially funded by a predoctoral fellowship from the Rett Syndrome Research Trust and Autism Science Foundation (#16–003). R.G. is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC PGS-D), UC San Diego Kavli Innovative Research Grant (IRG), Frontiers for Innovation Scholars Program fellowship, and a Katzin Prize. B.V. is supported by the Whitehall Foundation, a Sloan Research Fellowship, and the National Science Foundation under grant BCS-1736028.
Author contributions
ARM conceptualized the study. S.M.M. and A.R.M. designed all experiments. S.M.M. managed the animal colony, genotyping, and all animal behavior experiments. SW analyzed mouse open field data. S.M.M. performed and analyzed immunohistochemistry and immunocytochemistry. S.M.M. performed and analyzed MEA experiments. R.G. and B.V. analyzed MEA data. A.S. performed and analyzed calcium imaging experiments. Y.A., J.S.S. and J.S. assisted with in vitro assays. D.L. and H.S. performed and analyzed EEG experiments. J.E. and J.P.L. performed and analyzed MRI experiments. J.S., T.D.T. and C.K.G. analyzed RNA-Seq data. S.M.M. and A.R.M. wrote the original manuscript draft. C.K.G., A.R.M., and S.M.M. reviewed and edited the final manuscript draft.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflict of interest
Alysson R. Muotri, PhD, is a co-founder and has equity interest in TISMOO, a company dedicated to genetic analysis focusing on therapeutic applications customized for autism spectrum disorder and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. The remaining authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-018-0344-y).
References
- 1.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 2.Osipovich AB, Gangula R, Vianna PG, Magnuson MA. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development. 2016;143:4595–4607. doi: 10.1242/dev.141465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes, I. R. et al. Genetic variations on SETD5 underlying autistic conditions. Dev. Neurobiol. 78, 500–518 (2018). [DOI] [PubMed]
- 4.Grozeva D, et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am. J. Hum. Genet. 2014;94:618–624. doi: 10.1016/j.ajhg.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuechler A, et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur. J. Hum. Genet. 2015;23:753–760. doi: 10.1038/ejhg.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoner R, et al. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J. Neuropathol. Exp. Neurol. 2005;64:537–544. doi: 10.1093/jnen/64.6.537. [DOI] [PubMed] [Google Scholar]
- 10.Chailangkarn T, et al. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536:338–343. doi: 10.1038/nature19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaudoin GM, 3rd, et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 2012;7:1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- 12.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guyenet, S. J. et al. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp. (2010). http://www.jove.com/index/Details.stp?ID=1787, 10.3791/1787. [DOI] [PMC free article] [PubMed]
- 14.Katayama Y, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- 15.Crawley JN. Mouse behavioral assays relevant to the symptoms of autism. Brain. Pathol. 2007;17:448–459. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, M., Silverman, J. L. & Crawley, J. N. Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci.Chapter 8: Unit 8 26 (2011). [DOI] [PMC free article] [PubMed]
- 17.Gompers AL, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCabe AK, Chisholm SL, Picken-Bahrey HL, Moody WJ. The self-regulating nature of spontaneous synchronized activity in developing mouse cortical neurones. J. Physiol. 2006;577(Pt 1):155–167. doi: 10.1113/jphysiol.2006.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning JR, Jacobs J, Fried I, Kahana MJ. Broadband shifts in local field potential power spectra are correlated with single-neuron spiking in humans. J. Neurosci. 2009;29:13613–13620. doi: 10.1523/JNEUROSCI.2041-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gleichmann M, Mattson MP. Neuronal calcium homeostasis and dysregulation. Antioxid. Redox Signal. 2011;14:1261–1273. doi: 10.1089/ars.2010.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmunk G, Gargus JJ. Channelopathy pathogenesis in autism spectrum disorders. Front. Genet. 2013;4:222. doi: 10.3389/fgene.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinstry SU, et al. Huntingtin is required for normal excitatory synapse development in cortical and striatal circuits. J. Neurosci. 2014;34:9455–9472. doi: 10.1523/JNEUROSCI.4699-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellegood J, et al. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol. Psychiatry. 2015;20:118–125. doi: 10.1038/mp.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang DY, Beam D, Pelphrey KA, Abdullahi S, Jou RJ. Cortical morphological markers in children with autism: a structural magnetic resonance imaging study of thickness, area, volume, and gyrification. Mol. Autism. 2016;7:11. doi: 10.1186/s13229-016-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar JC, et al. Auditory encoding abnormalities in children with autism spectrum disorder suggest delayed development of auditory cortex. Mol. Autism. 2015;6:69. doi: 10.1186/s13229-015-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawa T, et al. Dysfunction of orbitofrontal and dorsolateral prefrontal cortices in children and adolescents with high-functioning pervasive developmental disorders. Ann. Gen. Psychiatry. 2013;12:31. doi: 10.1186/1744-859X-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Jiao J. Molecular biomarkers for embryonic and adult neural stem cell and neurogenesis. Biomed. Res. Int. 2015;2015:727542. doi: 10.1155/2015/727542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldiri I, et al. The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron. 2017;94:550–568 e510. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Science AIfB. Allen Developing Brain Reference Atlas, Allen Institute, Seattle, WA (2008). http://developingmouse.brain-map.org/static/atlas.
- 33.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl Acad. Sci. USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comery TA, et al. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl Acad. Sci. USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olde Loohuis NFM, et al. Altered expression of circadian rhythm and extracellular matrix genes in the medial prefrontal cortex of a valproic acid rat model of autism. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;77:128–132. doi: 10.1016/j.pnpbp.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J. Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang YT, Mason JO, Price DJ. Lateral cortical Cdca7 expression levels are regulated by Pax6 and influence the production of intermediate progenitors. Bmc. Neurosci. 2017;18:47. doi: 10.1186/s12868-017-0365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin HH, et al. Neuronatin promotes neural lineage in ESCs via Ca(2+) signaling. Stem Cells. 2010;28:1950–1960. doi: 10.1002/stem.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfurr S, et al. The E2A splice variant E47 regulates the differentiation of projection neurons viap57(KIP2) during cortical development. Development. 2017;144:3917–3931. doi: 10.1242/dev.145698. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, et al. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J. Cell. Mol. Med. 2016;20:2102–2110. doi: 10.1111/jcmm.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rooij, D. et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD working group. Am. J. Psychiatry175, 359–369 (2017). [DOI] [PMC free article] [PubMed]
- 43.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peier AM, et al. Over)correction of FMR1 deficiency with YAC transgenics: behavioral and physical features. Hum. Mol. Genet. 2000;9:1145–1159. doi: 10.1093/hmg/9.8.1145. [DOI] [PubMed] [Google Scholar]
- 45.Dachtler J, et al. Deletion of alpha-neurexin II results in autism-related behaviors in mice. Transl. Psychiatry. 2014;4:e484. doi: 10.1038/tp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiramoto T, et al. Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum. Mol. Genet. 2011;20:4775–4785. doi: 10.1093/hmg/ddr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.