Abstract
The pine wood nematode (PWN), Bursaphelenchus xylophilus, is the pathogen of pine wilt disease (PWD), resulting in huge losses in pine forests. However, its pathogenic mechanism remains unclear. The cathepsin L-like cysteine proteinase (CPL) genes are multifunctional genes related to the parasitic abilities of plant-parasitic nematodes, but their functions in PWN remain unclear. We cloned three cpl genes of PWN (Bx-cpls) by rapid amplification of cDNA ends (RACE) and analyzed their characteristics using bioinformatic methods. The tissue specificity of cpl gene of PWN (Bx-cpl) was studied using in situ mRNA hybridization (ISH). The functions of Bx-cpls in development and pathogenicity were investigated using real-time quantitative PCR (qPCR) and RNA interference (RNAi). The results showed that the full-length cDNAs of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 were 1163 bp, 1305 bp, and 1302 bp, respectively. Bx-cpls could accumulate specifically in the egg, intestine, and genital system of PWN. During different developmental stages of PWN, the expression of Bx-cpls in the egg stage was highest. After infection, the expression levels of Bx-cpls increased and reached their highest at the initial stage of PWD, then declined gradually. The silencing of Bx-cpl could reduce the feeding, reproduction, and pathogenicity of PWN. These results revealed that Bx-cpls play multiple roles in the development and pathogenic processes of PWN.
Keywords: Bursaphelenchus xylophilus, cathepsin L, gene expression, development, pathogenicity
1. Introduction
The pine wood nematode (PWN), Bursaphelenchus xylophilus, is the causal agent of pine wilt disease (PWD). It has been detected in North America (USA, Canada, and Mexico) [1,2], East Asia (Japan, China, and Korea) [3,4,5], Europe (Portugal and Spain) [6,7], and Nigeria [8]. The disease has been unquestionably a major threat to forest ecosystems worldwide and has caused great losses in China. However, the pathogenic mechanism of B. xylophilus remains unclear.
With the development of biotechnology, the expressed sequence tags (ESTs), genome, transcriptome, and secretome of B. xylophilus have been analyzed, highlighting several groups of genes putatively related to its pathogenicity [9,10,11,12,13]. Cellulase genes [14,15], pectatelyase genes [16,17], expansin-like genes [18,19], the venom allergen-like protein gene [20], and cytochrome P450 genes [21] have been studied and identified as pathogenesis-related genes. The functions of other putative pathogenesis-related genes of B. xylophilus still need to be identified.
It is believed that peptidases are essential for parasite development and in the most critical situations of parasite–host interactions. Peptidases comprise a large class of hydrolytic enzymes in parasites [22]. Of these, the cysteine peptidases are the class that covers virtually all functions that involve peptidases in parasitic helminths (including trematodes, cestodes, and nematode parasites) [23]. Cathepsin L is a type of cysteine peptidase belonging to the papain family and has been comprehensively studied in many parasitic helminths [24]. In free-living and parasite nematodes of humans and animals, the cathepsin L proteinases are involved in pivotal functions, such as tissue penetration, nutrition, immune evasion, and eggshell formation, though little is known of their precise functions [25]. As with animal parasite counterparts, nematodes that infect plants may require proteinases for egg hatching, larval molting, tissue penetration, and feeding. Urwin et al. [26] were the first team to clone a cathepsin L-like proteinase (CPL) from Heterodera glycines. To date, a number of cpl genes from plant parasitic nematodes including Bursaphelenchus, Globodera, Heterodera, Meloidogyne, and Rotylenchulus have been cloned, but their functions are seldom reported formally. The cpl gene of Meloidogyne incognita (Mi-cpl-1) encodes a digestive enzyme which is consistent with feeding [27,28]. In addition, Mi-cpl-1 can affect M. incognita development and play a crucial role in plant–nematode interactions [28,29,30]. The cpl gene in M. hispanica was also identified and characterized as a parasitism gene [31]. In B. xylophilus, two cpl genes (ACH69776.1, ACH56225.1) have been cloned, but their functions have not yet been investigated. In this study, the full-length cDNA of three novel cpl genes—Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3—were cloned using 3′ and 5′ rapid amplification of cDNA ends (RACE). The expressions of Bx-cpls in B. xylophilus at different developmental and pathogenic stages associated with PWD were analyzed by qPCR. The roles of Bx-cpls in reproduction and pathogenicity were verified through RNA interference (RNAi). These results provide useful information to better understand the functions of cpls in B. xylophilus and to elucidate the molecular pathogenic mechanism.
2. Results
2.1. Cloning and Sequence Analysis of Three Cathepsin L-Like Cysteine Proteinase Genes from B. xylophilus
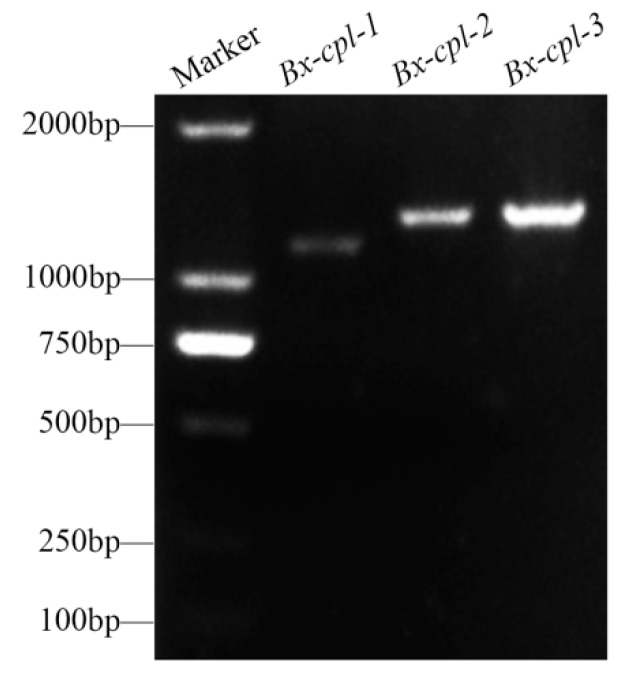
The complete nucleotide sequence of Bx-cpl-1 had 1163 bp (Figure 1), including an 18 bp 5′ untranslated region (UTR), a 1074 bp open reading frame (ORF), and a 71 bp 3′ UTR. It encodes a protein of 357 amino acid residues (Figure S1A). The full-length cDNA of Bx-cpl-2 had 1305 bp (Figure 1), comprising a 48 bp 5′ UTR, an 1185 bp ORF encoding 394 amino acid residues, and a 72 bp 3′ UTR (Figure S1B). The full-length cDNA of Bx-cpl-3 was 1302 bp (Figure 1), including a 51 bp 5′ UTR, a 63 bp 3′ UTR, and an 1188 bp ORF encoding 395 amino acids (Figure S1C). Compared to the genome data available on WormBase Parasite (BioProject PRJEA64437), the genomic locations of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 were at scaffold01141 191,468 to 192,800 with three introns, scaffold00813 265,318 to 266,698 with two introns, and scaffold01147 920,147 to 921,523 with two introns, respectively (Figure S2).
Figure 1.
Bands of Bx-cpls full-length cDNA sequences after gel electrophoresis.
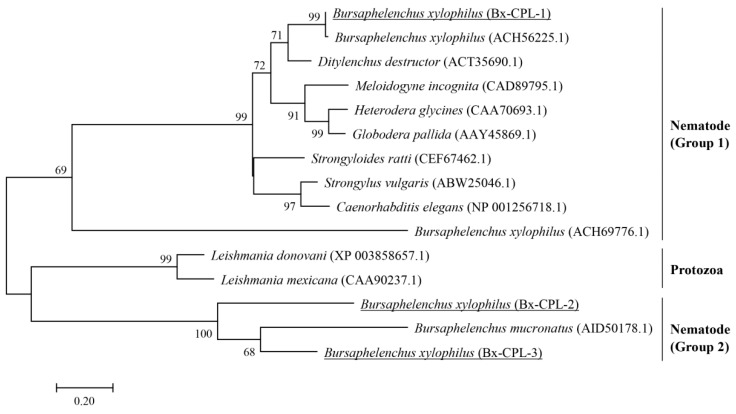
The results of Blastp showed that some CPL proteins have a relatively high level of identity with the CPLs of B. xylophilus. On this basis, amino acid sequences from Nematode and Protozoa which showed relatively high homology with the predicted amino acid sequences from B. xylophilus were selected and downloaded from NCBI. The phylogenetic tree was constructed by the maximum likelihood method with a WAG model with gamma-distributed rates based on the amino acid sequences of CPL proteins (Figure 2). Three CPL proteins of B. xylophilus were divided into two nematode groups. The Bx-cpl-1 deduced protein, Bx-CPL-1, is closely related to plant parasitic nematodes Ditylenchus destructor, Meloidogyne incognita, Heterodera glycines, Globodera pallida, and B. xylophilus in particular (ACH56225.1). However, the Bx-cpl-2 and Bx-cpl-3 deduced proteins, Bx-CPL-2 and Bx-CPL-3, were highly linked to B. mucronatus (AID50178.1) and even the CPLs in Protozoa, rather than the other CPLs in Nematode (Figure 2).
Figure 2.
Phylogenetic relationships of cathepsin L-like cysteine proteinases (CPLs). The phylogram was constructed based on amino acid sequences to determine the evolutionary relationships among 15 CPL proteins from different species using MEGA 7. The numbers below the branches indicate the bootstrap values, which were calculated from 1000 replicates. The GenBank accession numbers of the sequences are in brackets. B. xylophilus CPLs (Bx-CPL-1, Bx-CPL-2 and Bx-CPL-3) are underlined. Distance scale = 0.2.
2.2. Localization of Bx-cpl in B. xylophilus
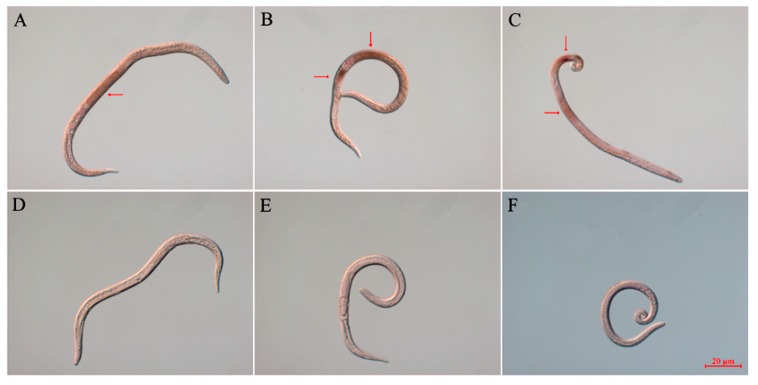
In situ hybridization (ISH) was used to analyze the tissue specificity of Bx-cpl transcription. The localizations of the three Bx-cpls were similar. The digoxigenin (DIG)-labeled antisense RNA probe of Bx-cpl generated clear signals in the intestine and egg of females (Figure 3A,B), and intestine and seminal vesicle of males of B. xylophilus (Figure 3C). No signals were observed in the control group with the sense probes.
Figure 3.
Localizations of Bx-cpls mRNA by in situ hybridization (ISH). Bx-cpl was expressed in PWNs in the intestine of females (A); the intestine and egg of females (B); and the intestine and seminal vesicle of males (C). The control groups showed no signals (D–F). The red arrows point to the hybridization signals. The scale bars are 20 µm.
2.3. Expression of Bx-cpl at PWN Developmental Stages
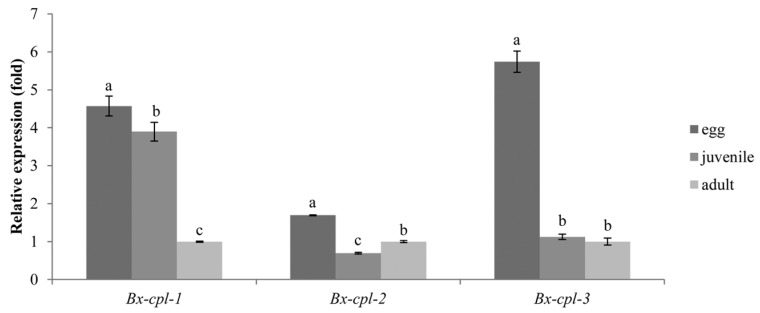
All Bx-cpls showed relatively high transcript levels in the egg stage. The Bx-cpl-1 expression was significantly lower in adults than in juveniles (p < 0.05), and Bx-cpl-2 expression was the opposite. There was no significant difference in Bx-cpl-3 expression between juveniles and adults (p > 0.05) (Figure 4).
Figure 4.
Relative expression levels of Bx-cpls at different developmental stages of B. xylophilus. The bars indicate standard errors, and different letters indicate significant differences (p < 0.05) among the different nematode stages (egg, juvenile, and adult).
2.4. Expression of Bx-cpl at PWD Development Stages
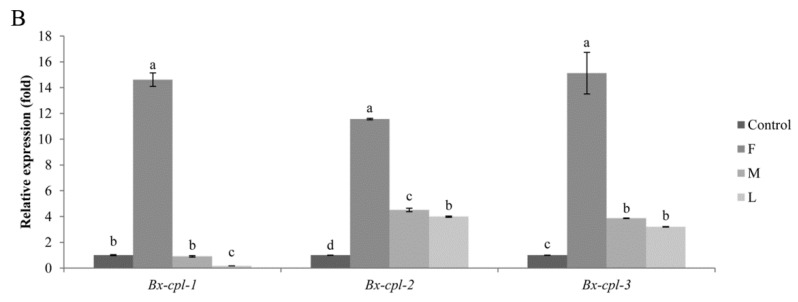
After infection of pine seedlings with B. xylophilus, all three Bx-cpls were found to be upregulated and reached the highest expression level at the first stage of PWD. Then, their expression declined and reached its lowest level at the late stage (Figure 5). These results indicate that the three Bx-cpls may play a similar role in PWD development, essentially at the early stages of PWD.
Figure 5.
Symptoms in P. massoniana after inoculation with nematodes: (A) First stage of pine wilt disease (PWD) (F), middle stage of PWD (M), and last stage of PWD (L). Pines inoculated with ddH2O (1); Pines inoculated with B. xylophilus (2). Relative expression levels of Bx-cpls at PWD development stages (B). The bars indicate standard errors, and different letters indicate significant differences (p < 0.05).
2.5. Detection of RNAi Efficiency
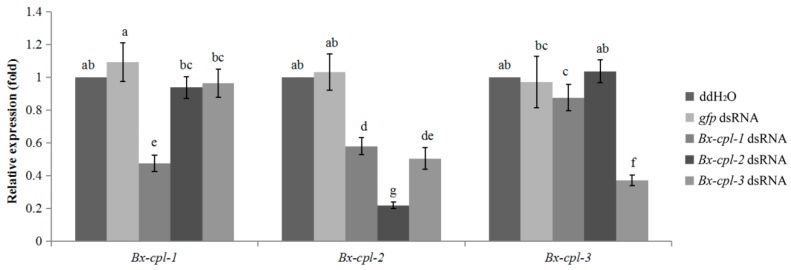
There were significant differences between each Bx-cpl transcript level in nematodes treated with the corresponding double-stranded RNA (dsRNA) and nematodes treated with ddH2O or green fluorescent protein gene (gfp) dsRNA (controls). The transcripts of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 decreased (p < 0.05) to 47.4, 21.8, and 37.0%, respectively, compared with in the nematodes treated with ddH2O (Figure 6). This showed that Bx-cpls expression was reduced by soaking the nematodes with the corresponding Bx-cpl dsRNA. In addition, dsRNA of Bx-cpl-1 and Bx-cpl-3 also targeted Bx-cpl-2 for degradation.
Figure 6.
Relative expression levels of Bx-cpls after treatment with Bx-cpl double-stranded RNA (dsRNA). The bars indicate standard errors, and different letters indicate significant differences (p < 0.05) among treatments: no dsRNA control (ddH2O), green fluorescent protein gene (gfp) dsRNA control, and each Bx-cpl dsRNA.
2.6. Feeding and Reproduction of B. xylophilus after RNAi
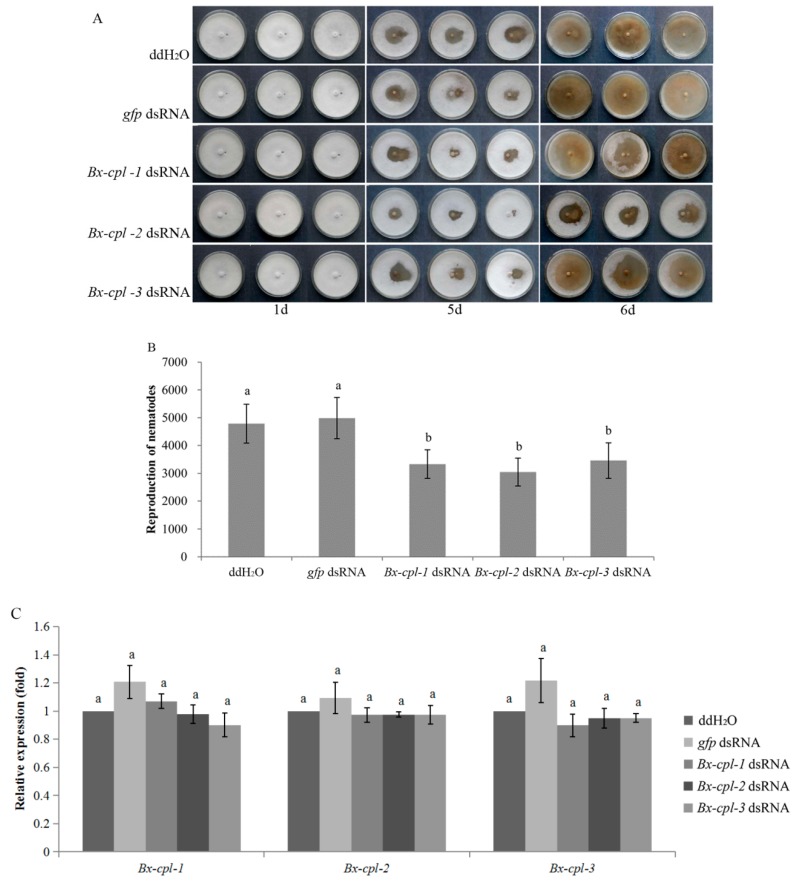
There was no significant difference in the feeding status between each treatment and the controls until Day 5 of nematode culture on Botrytis cinerea. The feeding areas of nematodes treated with ddH2O and gfp dsRNA were larger than those of nematodes treated with each Bx-cpl dsRNA (Figure 7A). At Day 6, the number of nematodes recovered from the culture plates was determined. The PWNs treated with Bx-cpl dsRNA were significantly fewer than those treated with ddH2O and gfp dsRNA (p < 0.05), and there was no significant difference (p > 0.05) between these two control treatments. Also, no significant differences were found between each Bx-cpl dsRNA treatment (Figure 7B). At this time, the expression levels of all three cpl genes under each of the five treatments (ddH2O and dsRNA (gfp, Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3)) were detected. There was no significant difference among all the treatments (p > 0.05). These results indicated a deleterious effect of silencing of Bx-cpl on the development of B. xylophilus, but one which is limited by time.
Figure 7.
Effects of RNA interference (RNAi) on feeding and reproduction of B. xylophilus. RNAi-treated B. xylophilus after cultivation on B. cinerea (A); Total B. xylophilus population recovered from B. cinerea plates six days after treatment with ddH2O and dsRNA (gfp, Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3) (B); Relative expression levels of Bx-cpls after cultivation on B. cinerea for six days (C). The bars indicate standard errors between replicates, and different letters indicate significant differences (p < 0.05) among treatments.
2.7. Pathogenicity of B. xylophilus after RNAi
Five days after inoculation, the pines with nematodes treated with ddH2O and gfp dsRNA showed clear symptoms. Their infection rates were 25% and 50%, and the disease severity index (DSI) values were 6.25 and 12.25, respectively. At this timepoint, no visible symptoms were registered for pine trees inoculated with nematodes treated with Bx-cpl dsRNA. Eight days after inoculation, the pines inoculated with nematodes treated with Bx-cpl-1 and Bx-cpl-3 dsRNA showed symptoms with infection rates of 25% and DSI values of 6.25 (Table 1). The pines inoculated with nematodes treated with Bx-cpl-2 dsRNA presented leaf browning only at the ninth day after inoculation. At Day 20, most of the pines inoculated with PWNs developed symptoms (Figure 8). Thirty-five days after inoculation, the infection rates were all 100%, but the DSI values of the Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 dsRNA treatments (62.5, 50, 56.25) were lower than those of the ddH2O and gfp dsRNA treatments (93.75, 100). These results showed that the pathogenicity of B. xylophilus decreased after treatment with Bx-cpl dsRNA.
Table 1.
Symptoms of Pinus massoniana caused by B. xylophilus treated with dsRNA.
| Treatment | Infection Rates (%) | Disease Severity Index (DSI) | ||||||
|---|---|---|---|---|---|---|---|---|
| 5th Day | 8th Day | 20th Day | 35th Day | 5th Day | 8th Day | 20th Day | 35th Day | |
| ddH2O | 25 | 50 | 75 | 100 | 6.25 | 25 | 31.25 | 93.75 |
| gfp dsRNA | 50 | 100 | 100 | 100 | 12.5 | 37.5 | 68.75 | 100 |
| Bx-cpl-1 dsRNA | 0 | 25 | 75 | 100 | 0 | 6.25 | 18.75 | 62.5 |
| Bx-cpl-2 dsRNA | 0 | 0 | 50 | 100 | 0 | 0 | 12.5 | 50 |
| Bx-cpl-3 dsRNA | 0 | 25 | 50 | 100 | 0 | 6.25 | 12.5 | 56.25 |
Figure 8.
Symptoms in P. massoniana after inoculation with nematodes. Symptoms 0 days after inoculation (A); Symptoms 8 days after inoculation (B); Symptoms 20 days after inoculation (C); Symptoms 35 days after inoculation (D). Pines inoculated with ddH2O (1); B. xylophilus soaked in ddH2O (2); B. xylophilus gfp dsRNA (3); B. xylophilus Bx-cpl-1 dsRNA (4); B. xylophilus Bx-cpl-2 dsRNA (5); B. xylophilus Bx-cpl-3 dsRNA (6).
3. Discussion
Cathepsin L-like cysteine proteinase (CPL) is a protease widely distributed in tissues and cells. In many parasitic nematodes, CPL plays an important role in molting, individual development, invasion, feeding on host tissues, and evasion of innate host defenses [28,32,33,34]. However, the roles of CPL in B. xylophilus remain unknown.
In this study, the full-length cDNA of three Bx-cpls were cloned, and their amino acid sequences were deduced. Homology analysis showed that the Bx-cpl-1 deduced protein, Bx-CPL-1, has a close phylogenetic relationship with the CPLs of plant parasitic nematodes such as D. destructor, M. incognita, and H. glycines. Wang et al. [35] analyzed the homology between the deduced protein of a cpl sequence from D. destructor (ACT35690) and the CPL of B. xylophilus (ACH56225.1) and found that the identities were highly similar. The CPL of M. incognita also had a high homology with Bx-CPL-1, which plays a crucial role in plant–nematode interaction [30]. The Bx-cpl-2 and Bx-cpl-3 deduced proteins (Bx-CPL-2 and Bx-CPL-3) have a close phylogenetic relationship with a CPL of B. mucronatus (AID50178.1), which may be related to infection of B. mucronatus [36].
ISH enables the investigation of gene expression patterns and gene functions in nematodes [28,37,38]. Hashmi et al. [37] reported that the CPL is widely expressed in the head region, intestines, hypodermal cells, and eggshells of Caenorhabditis elegans. However, in plant parasitic nematode M. incognita, Mi-cpl-1 was only expressed in the intestinal cells of M. incognita [27,28]. In this study, the localizations of Bx-cpl-1, Bx-cpl-2, Bx-cpl-3 were all in the intestine and egg of the female PWN and in the intestine and seminal vesicle of the male PWN. This suggests that the expressions of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 are similar. The Bx-CPL protein might be involved in the digestive and reproductive processes of B. xylophilus.
The CPL could regulate the nematode’s development [39]. In this study, the three Bx-cpls were differently expressed in different developmental stages of B. xylophilus, and the expressions in eggs were relatively higher than those in juveniles and adults. Hashmi et al. [37] demonstrated that the CPL was essential for the embryogenesis and development of C. elegans. This suggests that the Bx-cpls might play a role in the development of B. xylophilus, especially in embryogenesis.
The relative expression levels of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 at PWD development stages were also investigated. The transcript levels of the three Bx-cpls from P. massoniana (except the transcript level of Bx-cpl-1 at the last stage) were higher than in the nematodes cultured on B. cinerea. This characteristic is similar to many pathogenesis-related genes of B. xylophilus, such as pectate lyase genes, cytochrome P450 (CYP450) genes, UDP-glucuronosyltrans-ferase (UGT) genes, and ATP-binding cassette (ABC) transporter genes, which were expressed more highly when B. xylophilus infected P. thunbergii than when it was cultured on B. cinerea [40]. Kang et al. [41] constructed subtractive expressed sequence tag (EST) libraries that were specific to the dispersal 4th larval stage (D4S) and the pine-grown propagative mixed stage (PGPS) and found that cysteine protease was highly specific to PGPS compared to D4S. The relative expression levels of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 were highest at the first stage but declined with the development of the PWD. This suggests that Bx-cpls might be associated with the parasitic biology of B. xylophilus during its propagation within the host pine tree, especially at the first stage of PWD.
RNAi is a means by which double-stranded RNA (dsRNA) induces sequence-specific post-transcriptional gene silencing [42]. It is a very powerful tool for examining the functions of genes in plant nematodes and other organisms [28,32,43,44]. RNAi-induced gene silencing has previously been achieved in B. xylophilus in vitro [21,38,45,46,47]. In this study, the expression levels of Bx-cpls treated with Bx-cpl dsRNA significantly decreased compared to those of the control groups, indicating that Bx-cpl genes could be silenced. In many parasitic nematodes, CPLs have potential roles in invasion and feeding on host tissues, molting, development, and parasitism [27,28,30,37,48,49]. Our results showed that the feeding of PWN weakened, reproduction was reduced, and pathogenicity was lower after silencing Bx-cpl-1, Bx-cpl-2, or Bx-cpl-3, respectively. This suggests that Bx-cpls could regulate the nematodes’ reproduction and pathogenicity.
4. Materials and Methods
4.1. Nematode Culture and Collection
Bursaphelenchus xylophilus AMA3 isolated from infected P. thunbergii in Maanshan, Anhui, China was provided by the Jiangsu Key Laboratory for Prevention and Management of Invasive Species, Nanjing Forestry University.
The PWNs at different developmental stages were collected according to the method described by Shinya et al. [50] with some modifications. The nematodes were cultured on potato dextrose agar (PDA) covered with Botrytis cinerea at 25 °C for 4–5 days and isolated with Baermann funnels. The nematodes were washed three times with distilled water and collected by centrifugation at 3500 rpm for 3 min. Approximately 5000 nematodes were placed in a sterilized plate (3 cm diameter) to lay eggs for 4–6 h at 25 °C under aseptic conditions. The eggs were collected after the nematodes were discarded by sterile water washing. The juveniles, including the second-juveniles (J2), the third-juveniles (J3), and the forth-juveniles (J4), were obtained after the eggs were cultured on a PDA plate containing B. cinerea for 30–48 h. The adults, including male and female nematodes, were obtained after the eggs were cultured for 84 h. The nematodes at different developmental stages were identified under a microscope (Leica DM500, Leica Microsystems, Heerbrugg, Switzerland). Then, the nematodes in the same developmental stage were washed three times with distilled water and collected by centrifugation at 3500 rpm for 3 min. The collected nematodes were immediately frozen in liquid nitrogen and stored at −80 °C in a 1.5 mL centrifuge tube for subsequent RNA extraction.
The PWNs at different PWD development stages were collected according to the method described by Ding et al. [51]. The seedlings of P. massoniana (2 years old) were disinfected with 75% ethyl alcohol by spraying. Afterwards, 0.5 mL suspensions (about 10,000 mixed-stage nematodes) were pipetted into cutting wounds in P. massoniana. Sterile water was used as the control. Then the wounds on P. massoniana were sealed by Parafilm. PWNs were collected from three stages based on the corresponding PWD symptoms and inoculation times. In the first stage (F), the tips of the pine needles began to turn brown after pine trees were infected with PWNs. Next, in the middle stage (M), half of the needles on the pine trees turned brown. In the last stage (L), the pine needles were completely brown. PWNs cultured on B. cinerea served as a control. The nematodes were extracted with Baermann funnels and washed three times with distilled water. Then, they were collected by centrifugation at 3500 rpm for 3 min, immediately frozen in liquid nitrogen, and stored at −80 °C in a 1.5 mL centrifuge tube for subsequent RNA extraction.
4.2. RNA Extraction, PCR Amplification of Bx-cpls, and Phylogenetic Analysis
Total RNA was extracted from the nematodes at each developmental stage and mixed stages using Trizol reagent (Invitrogen, Waltham, MA, USA), measured by ultraviolet absorbance at A260/280 (Eppendorf AG 22331, Hamburg, Germany), and examined by electrophoresis on a 1% agarose gel. The cDNA was synthesized using the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix according to the manufacturer’s instructions (TransGen Biotech, Beijing, China). The full-length cDNA sequences of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 were amplified using the 3′-Full RACE Core Set with the PrimeScript™ RTase kit (TaKaRa Biotechnology, Dalian, China) and 5′-Full RACE Kit with TAP (TaKaRa Biotechnology, Dalian, China). Gene-specific primers were used as follows: Bx-cpl-1: GSP1-1 (3′-Full RACE first round of PCR), GSP1-2 (5′-Full RACE first round of PCR), and GSP1-3 (5′-Full RACE second round of PCR); Bx-cpl-2: GSP2-1 (3′-Full RACE first round of PCR) and GSP2-2 (5′-Full RACE first round of PCR); Bx-cpl-3: GSP3-1 (3′-Full RACE first round of PCR), GSP3-2 (3′-Full RACE second round of PCR), GSP3-3 (5′-Full RACE first round of PCR), and GSP3-4 (5′-Full RACE second round of PCR) (Table 2). They were designed for 3′ and 5′ RACE amplification based on three partially known sequences of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 which were obtained from the RNA sequencing results [52]. The PCR product was purified, ligated into the vector pEASY-T1 (TransGen Biotech, Beijing, China), and transformed into Escherichia coli Trans1-T1 (E. coli) competent cells (TransGen Biotech, Beijing, China). The E. coli was then incubated overnight at 37 °C on Luria-Bertani (LB) plates containing ampicillin. The positive transformants were analyzed by PCR using primers M13F (-47) and M13R (-48) (Table 2). Once the correct clone was identified, the fresh bacterial suspension was submitted to the Nanjing Genscript sequencing company (Nanjing, China) for sequence analysis. The full-length cDNA sequences of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 from B. xylophilus were submitted to GenBank and assigned the accession numbers MG923677, MG923678, and MG923679. A reference and comparison to the genome data available on WormBase Parasite (BioProject PRJEA64437) were performed using blastn (https://parasite.wormbase.org/Multi/Tools/Blast?db=core) and DNAMAN software (https://www.lynnon.com/index.html). Amino acid sequences of homologous Bx-CPL-1, Bx-CPL-2, and Bx-CPL-3 proteins from other species were obtained from NCBI using blastp. Multiple sequence alignment of deduced protein sequences was carried out with ClustalW in MEGA 7 (https://www.megasoftware.net/) [53]. WAG model with gamma-distributed rates (WAG+G) resulted as the best model by Find Best-fit Substitution Model in MEGA 7. Phylogenetic relationships among the CPLs were inferred by using the Maximum Likelihood (ML) method with a WAG+G model.
Table 2.
Polymerase chain reaction (PCR) primers.
| Name of Primers | Sequence (5′–3′) |
|---|---|
| cDNA Cloning of Three Cathepsin L-like Cysteine Proteinase Genes | |
| 3′ RACE (rapid amplification of cDNA ends) outer primer | TACCGTCGTTCCACTAGTGATTT |
| 3′ RACE inner primer | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| GSP (gene specific primer) 1-1 | GCAATGGTGGACTTATGGAC |
| GSP2-1 | AATCCAAGAGCCCCGTTATC |
| GSP3-1 | GCACCTACCGAAGCCGATACTA |
| GSP3-2 | CCACTCCAAGACTACCAAGG |
| 5′RACE outer primer | CATGGCTACATGCTGACAGCCTA |
| 5′RACE inner primer | CGCGGATCCACAGCCTACTGATGATCAGTCGATG |
| GSP1-2 | CTTGACGATCCAGTAGTCGC |
| GSP1-3 | CTCGCCATTTGGTCGCATTT |
| GSP2-2 | GGTTCTATCGCCGACATTCT |
| GSP3-3 | AACCAAAGTGTAGCCCCAAT |
| GSP3-4 | TGACCAAAGCGTTGCGAAGT |
| M13F(−47) | CGCCAGGGTTTTCCCAGTCACGAC |
| M13R(−48) | AGCGGATAACAATTTCACACAGGA |
| Preparation of Template DNA for ISH | |
| I-Bx-cpl-1-F | CCTTTCGCTGAATACCGTCGTCTTA |
| I-Bx-cpl-1-R | TGATGACTCAAGCCAGCGGATAACT |
| I-Bx-cpl-1-T7-F | TAATACGACTCACTATAGGGCCTTTCGCTGAATACCGTCGTCTTA |
| I-Bx-cpl-1-T7-R | TAATACGACTCACTATAGGGTGATGACTCAAGCCAGCGGATAACT |
| I-Bx-cpl-2-F | GCTGTGGATGTTGCTACGCTTTTGC |
| I-Bx-cpl-2-R | GCTTCTCCGTAGTCCTCTCCCCATT |
| I-Bx-cpl-2-T7-F | TAATACGACTCACTATAGGGGCTGTGGATGTTGCTACGCTTTTGC |
| I-Bx-cpl-2-T7-R | TAATACGACTCACTATAGGGGCTTCTCCGTAGTCCTCTCCCCATT |
| I-Bx-cpl-3-F | ACAGCAGTGCCAAGCCCGCTCAAAT |
| I-Bx-cpl-3-R | GTGCTCGGGCATTGATGATTCCTCC |
| I-Bx-cpl-3-T7-F | TAATACGACTCACTATAGGGACAGCAGTGCCAAGCCCGCTCAAAT |
| I-Bx-cpl-3-T7-R | TAATACGACTCACTATAGGGGTGCTCGGGCATTGATGATTCCTCC |
| Preparation of Template DNA for dsRNA | |
| Bx-cpl-1-T7-F | GCCAGTCGTCATCACAAA |
| Bx-cpl-1-R | TGTTCCTCATCGGCTTCT |
| Bx-cpl-1-F | TAATACGACTCACTATAGGGGCCAGTCGTCATCACAAA |
| Bx-cpl-1-T7-R | TAATACGACTCACTATAGGGTGTTCCTCATCGGCTTCT |
| Bx-cpl-2-T7-F | TAATACGACTCACTATAGGGACTAGATCCCAGCGCCACT |
| Bx-cpl-2-R | AGCCAACAGTCACGACAGC |
| Bx-cpl-2-F | ACTAGATCCCAGCGCCACT |
| Bx-cpl-2-T7-R | TAATACGACTCACTATAGGGAGCCAACAGTCACGACAGC |
| Bx-cpl-3-T7-F | TAATACGACTCACTATAGGGAGAGCTTCACAGCAGTGCCAAG |
| Bx-cpl-3-R | GTTGAACCTGGTAACTATAGTC |
| Bx-cpl-3-F | GCTTCACAGCAGTGCCAAG |
| Bx-cpl-3-T7-R | TAATACGACTCACTATAGGGAGAGTTGAACCTGGTAACTATAGTC |
| gfp-T7-F | TAATACGACTCACTATAGGGAGACCATGGCCAACACTTGT |
| gfp-R | AGATAATCCCAGCAGCAGTT |
| gfp-F | AGACCATGGCCAACACTTGT |
| gfp-T7-R | TAATACGACTCACTATAGGGAGATAATCCCAGCAGCAGTT |
| Real Time PCR | |
| q-Bx-cpl-1-F | CCAGAAGCCGATGAGGAACA |
| q-Bx-cpl-1-R | CCAGTTTTGTAGAGTTGGAAGC |
| q-Bx-cpl-2-F | AGTCATCGCTGTAATCTGC |
| q-Bx-cpl-2-R | TTGTTGGTGCCATAAGTG |
| q-Bx-cpl-3-F | CTATAACGGAGTCACCTCCAT |
| q-Bx-cpl-3-R | TGCTCTTCACTGAGATCCAGT |
| Actin-F | GCAACACGGAGTTCGTTGTAGA |
| Actin-R | GTATCGTCACCAACTGGGATGA |
4.3. In Situ Hybridization (ISH)
The ISH probe templates were generated by PCR based on the full-length cDNA sequences of Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 with the specific primer pairs (Table 2). The DIG-labeled sense RNA probes and antisense RNA probes were synthesized from the PCR products of Bx-cpls using the DIG Northern Starter Kit (Roche Diagnostics, Mannheim, Germany) [54]. The nematodes were treated, and hybridizations were performed as described by De Boer et al. [55] using a DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Diagnostics, Mannheim, Germany). For the control group, DIG-labeled sense RNA probes were used. Finally, the nematodes were examined and photographed using a Zeiss Axio Image M2 microscope (Zeiss MicroImaging GmbH, Oberkochen, Germany).
4.4. Synthesis of Bx-cpl dsRNA and Interference
Double-stranded RNA (dsRNA) was synthesized using the MEGscript RNAi Kit (Ambion Inc., Austin, TX, USA) with the specific primers containing the T7 promoter (Table 2). The non-endogenous control dsRNA (the green fluorescent protein gene, gfp) was synthesized using the specific primers gfp-T7-F/gfp-R and gfp-F/gfp-T7-R (Table 2). The RNAi soaking method was performed following the process by Urwin et al. [32]. Approximately 3000 individuals (a mixture of juveniles and adults) of freshly cultured nematodes were soaked in dsRNA solution (800 ng/µL) after being washed with distilled water three times at 3500 rpm for 3 min, and then incubated at 180 rpm for 48 h at 20 °C. The nematodes soaked in the corresponding gfp dsRNA and ddH2O were used as controls. Each treatment had three replicates. Samples from each treatment were washed thoroughly with ddH2O several times after soaking and then used for additional experiments.
4.5. The qPCR and Expression Analysis of Bx-cpls
Expressions of Bx-cpls were analyzed using real-time quantitative PCR (qPCR). Total RNA was extracted from 3000 nematodes at each developmental stage and mixed stages using Trizol reagent (Invitrogen, Waltham, MA, USA). The RNA quantity and integrity were checked as previously described. The cDNA was synthesized using TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix following the manufacturer’s protocol (TransGen Biotech, Beijing, China). Specific primers were designed from the cDNA sequence of target genes using Primer Premier 5.0 (Table 2). The actin gene was amplified as a reference gene using the primers Actin-F/Actin-R (Table 2). The qPCR was performed on ABI Prism 7500 (Applied Biosystems, Foster City, CA, USA) using SYBR Green Master Mix (Vazyme, Nanjing, China). The initial data analysis was performed using ABI Prism 7500 software (https://www.thermofisher.com/cn/zh/home/technical-resources/software-downloads/applied-biosystems-7500-fast-real-time-pcr-system.html) and the 2−ΔΔCt method. All experiments were performed in triplicate with three biological replicates.
4.6. Analysis of Reproduction and Pathogenicity of B. xylophilus after RNAi
About 200 nematodes treated with Bx-cpl dsRNA were cultured on a PDA plate with B. cinerea at 25 °C for 6 days. The ddH2O and gfp dsRNA were used as controls. Each treatment had three replicates. The feeding of B. xylophilus was observed and photographed periodically. Subsequently, the nematodes were washed off the plates using a Baermann funnel. The reproduction rate of the nematodes was counted with an optical microscope (Leica DM500, Leica Microsystems, Heerbrugg, Switzerland). In order to determine the pathogenicity of B. xylophilus after RNAi, nearly 2000 nematodes soaked in Bx-cpl dsRNA, gfp dsRNA, or ddH2O without dsRNA were inoculated into each 4-year-old P. massoniana seedling. ddH2O without nematodes was used as an inoculation control. Each treatment contained four replicates. The inoculated seedlings were placed in a greenhouse. Photographs were taken regularly to record the infection state of the seedlings. PWD symptoms were evaluated and categorized on a scale from 0 to 4 [56]. The categories were as follows: 0 = all needles were green; 1 = 0%–25% of needles were discolored and turned yellow; 2 = 25%–50% of needles turned yellow; 3 = 50%–75% of needles turned yellow; and 4 = 75%–100% of needles turned yellow. The infection rates and the disease severity index (DSI) were calculated using the following formulae:
4.7. Statistical Analysis
All data are presented as the mean ± standard deviation (Mean ± SD). All parameters were calculated using Microsoft Excel. The statistical significance was determined using SPSS Statistics 17.0 software (IBM China Company Ltd., Beijing, China) with ANOVA and t-tests. The level of significance was p < 0.05.
5. Conclusions
Bx-cpl-1, Bx-cpl-2, and Bx-cpl-3 are three cathepsin L-like cysteine proteinase genes of B. xylophilus in different genomic locations. Their expressions and functions are similar. They are all involved in the feeding, digestion, development, reproduction, and parasitism of B. xylophilus. Silencing of Bx-cpl would result in decreasing the feeding ability, number of nematodes, and development of pine wilt disease. These results indicate that cathepsin L-like cysteine proteinase genes play a regulatory role in the development and pathogenicity of the pine wood nematode. This is beneficial to better understanding the molecular mechanisms of development and pathogenicity in B. xylophilus.
Acknowledgments
We are grateful to De-Wei Li, The Connecticut Agricultural Experiment Station, USA for reviewing the manuscript.
Supplementary Materials
Supplementary materials are available online http://www.mdpi.com/1422-0067/20/1/215/s1. Figure S1. Full-length cDNA sequences and deduced amino acid sequences of the Bx-cpls Bx-cpl-1 (A), Bx-cpl-2 (B), and Bx-cpl-3 (C). Figure S2: Comparison of Bx-cpls gene sequences to genome data of B. xylophilus. Bx-cpl-1 gene (A); Bx-cpl-2 gene (B); Bx-cpl-3 gene (C). The intron sequences are underlined.
Author Contributions
Conceptualization, Q.X. and X.-Q.W.; methodology, Q.X. and X.-Q.W.; validation, Q.X., X.-Q.W. and L.-N.D.; formal analysis, Q.X.; investigation, Q.X., W.-J.Z. and M.-M.W.; resources, X.-Q.W.; data curation, Q.X., W.-J.Z. and M.-M.W.; writing—original draft preparation, Q.X.; writing—review and editing, Q.X. and X.-Q.W.; visualization, Q.X.; supervision, X.-Q.W.; project administration, Q.X. and X.-Q.W.; funding acquisition, X.-Q.W.
Funding
This research was funded by the Jiangsu Provincial Agricultural Science and Technology Innovation Fund (CX (16) 1005), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Innovation Plan for Graduate Students of Jiangsu, China (KYZZ16_0315).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Shinya R., Morisaka H., Takeuchi Y., Futai K., Ueda M. Making headway in understanding pine wilt disease: What do we perceive in the postgenomic era? J. Biosci. Bioeng. 2013;116:1–8. doi: 10.1016/j.jbiosc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Dwinell L.D. First report of pinewood nematode (Bursaphelenchus xylophilus) in Mexico. Plant Dis. 1993;77:846A. doi: 10.1094/PD-77-0846A. [DOI] [Google Scholar]
- 3.Mamiya Y. History of pine wilt disease in Japan. J. Nematol. 1988;20:219–226. doi: 10.1002/jez.1402440122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K., Liang J., Yan D.H., Zhang X.Y. Research advances of pine wood nematode disease in China. World For. Res. 2010;23:59–63. doi: 10.13348/j.cnki.sjlyyj.2010.03.008. [DOI] [Google Scholar]
- 5.Yi C.K., Byun B.H., Park J.D., Yang S.I., Chang K.H. First finding of the pine wood nematode, Bursaphelenchus xylophilus (Steiner et Buhrer) Nickle and its insect vector in Korea. Res Rep For Res Inst Seoul. 1989;3:141–149. [Google Scholar]
- 6.Mota M.M., Braasch H., Bravo M.A., Penas A.C., Burgermeister W., Metge K., Sousa E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology. 1999;1:727–734. doi: 10.1163/156854199508757. [DOI] [Google Scholar]
- 7.Abelleira A., Picoaga A., Mansilla J.P., Aguin O. Detection of Bursaphelenchus xylophilus, causal agent of pine wilt disease on Pinus pinaster in northwestern Spain. Plant Dis. 2011;95:776. doi: 10.1094/PDIS-12-10-0902. [DOI] [PubMed] [Google Scholar]
- 8.Khan F.A., Gbadegesin R.A. On the occurrence of nematode induced pine wilt disease in Nigeria. Pak. J. Nematol. 1991;57:162–164. doi: 10.1037/0022-006X.57.1.162. [DOI] [Google Scholar]
- 9.Kikuchi T., Aikawa T., Kosaka H., Pritchard L., Ogura N., Jones J.T. Expressed sequence tag (EST) analysis of the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus. Mol. Biochem. Parasitol. 2007;155:9–17. doi: 10.1016/j.molbiopara.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi T., Cotton J.A., Dalzell J.J., Hasegawa K., Kanzaki N., McVeigh P., Takanashi T., Tsai I.J., Assefa S.A., Cock P.J., et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinya R., Morisaka H., Kikuchi T., Takeuchi Y., Ueda M., Futai K. Secretome Analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS ONE. 2013;8:e67377. doi: 10.1371/journal.pone.0067377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai I.J., Tanaka R., Kanzaki N., Akiba M., Yokoi T., Espada M., Jones J.T., Kikuchi T. Transcriptional and morphological changes in the transition from mycetophagous to phytophagous phase in the plant-parasitic nematode Bursaphelenchus xylophilus. Mol. Plant Pathol. 2016;17:77–83. doi: 10.1111/mpp.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso J., Anjo S., Fonseca L., Egas C., Manadas B., Abrantes I. Bursaphelenchus xylophilus and B. mucronatus secretomes: A comparative proteomic analysis. Sci. Rep. 2016;6:39007. doi: 10.1038/srep39007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi T., Jones J.T., Aikawa T., Kosaka H., Ogura N. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004;572:201–205. doi: 10.1016/j.febslet.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Fan Y., Zheng H., Du F., Zhang K.Q., Huang X., Wang L., Zhang M., Niu Q. Isolation and characterization of a novel endoglucanase from a Bursaphelenchus xylophilus metagenomic library. PLoS ONE. 2013;8:e82437. doi: 10.1371/journal.pone.0082437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi T., Shibuya H., Aikawa T., Jones J.T. Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 2006;19:280–287. doi: 10.1094/MPMI-19-0280. [DOI] [PubMed] [Google Scholar]
- 17.Lee D.W., Kang J.S., Jung C.S., Han H.R., Moon Y.S., Park S.J., Lee S.H., Koh Y.H. Identification and biochemical analysis of a novel pectate lyase 3 gene in Bursaphelenchus xylophilus. J. Asia-Pac. Entomol. 2013;16:335–342. doi: 10.1016/j.aspen.2013.04.016. [DOI] [Google Scholar]
- 18.Kikuchi T., Li H.M., Karim N., Kennedy M.W., Moens M., Jones J.T. Identification of putative expansin-like genes from the pine wood nematode, Bursaphelenchus xylophilus, and evolution of the expansin gene family within the nematoda. Nematology. 2009;11:355–364. doi: 10.1163/156854109X446953. [DOI] [Google Scholar]
- 19.Kim Y.H., Kim A.Y., Choi B.H., Han H.R., Koh Y.H. ExpansinB3 as a marker for detecting pine wood nematode-infected pine trees. J. Asia-Pac. Entomol. 2017;20:1228–1233. doi: 10.1016/j.aspen.2017.08.029. [DOI] [Google Scholar]
- 20.Lin S.F., Jian H., Zhao H.J., Yang D., Liu Q. Cloning and characterization of a venom allergen-like protein gene cluster from the pinewood nematode Bursaphelenchus xylophilus. Exp. Parasitol. 2011;127:440–447. doi: 10.1016/j.exppara.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Xu X.L., Wu X.Q., Ye J.R., Huang L. Molecular characterization and functional analysis of three pathogenesis-related cytochrome P450 genes from Bursaphelenchus xylophilus (Tylenchida, Aphelenchoidoidea) Int. J. Mol. Sci. 2015;16:5216–5234. doi: 10.3390/ijms16035216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhoads M.L., Fetterer R.H. Extracellular matrix: A tool for defining the extracorporeal function of parasite proteases. Parasitol. Today. 1997;13:119–122. doi: 10.1016/S0169-4758(96)40011-4. [DOI] [PubMed] [Google Scholar]
- 23.Malagón D., Benítez R., Kašný M., Adroher F.J. Peptidases in parasitic nematodes. A review. In: Erzinger G.S., editor. Parasites: Ecology, Diseases and Management. Nova Science Publishers Inc.; New York, NY, USA: 2013. pp. 61–102. [Google Scholar]
- 24.Sajid M., McKerrow J.H. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 2002;120:1–21. doi: 10.1016/S0166-6851(01)00438-8. [DOI] [PubMed] [Google Scholar]
- 25.Britton C., Murray L. A cathepsin L protease essential for Caenorhabditis elegans embryogenesis is functionally conserved in parasitic nematodes. Mol. Biochem. Parasitol. 2002;122:21–33. doi: 10.1016/S0166-6851(02)00066-X. [DOI] [PubMed] [Google Scholar]
- 26.Urwin P.E., Lilley C.J., McPherson M.J., Atkinson H.J. Characterization of two cDNAs encoding cysteine proteinases from the soybean cyst nematode Heterodera glycines. Parasitology. 1997;114:605–613. doi: 10.1016/S0166-6851(97)00116-3. [DOI] [PubMed] [Google Scholar]
- 27.Neveu C., Abad P., Castagnone-Sereno P. Molecular cloning and characterization of an intestinal cathepsin L protease from the plant-parasitic nematode Meloidogyne incognita. Physiol. Mol. Plant Pathol. 2003;63:159–165. doi: 10.1016/j.pmpp.2003.10.005. [DOI] [Google Scholar]
- 28.Shingles J., Lilley C.J., Atkinson H.J., Urwin P.E. Meloidogyne incognita: Molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp. Parasitol. 2007;115:114–120. doi: 10.1016/j.exppara.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Neveu C., Jaubert S., Abad P., Castagnone-Sereno P. A set of genes differentially expressed between avirulent and virulent Meloidogyne incognita near-isogenic lines encode secreted proteins. Mol. Plant Microbe Interact. 2003;16:1077. doi: 10.1094/MPMI.2003.16.12.1077. [DOI] [PubMed] [Google Scholar]
- 30.Dutta T.K., Papolu P.K., Banakar P., Choudhary D., Sirohi A., Rao U. Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 2015;6:260. doi: 10.3389/fmicb.2015.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duarte A., Maleita C., Tiago I., Curtis R., Abrantes I. Molecular characterization of putative parasitism genes in the plant-parasitic nematode Meloidogyne hispanica. J. Helminthol. 2016;90:28–38. doi: 10.1017/S0022149X1400073X. [DOI] [PubMed] [Google Scholar]
- 32.Urwin P.E., Lilley C.J., Atkinson H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 2002;15:747–752. doi: 10.1094/MPMI.2002.15.8.747. [DOI] [PubMed] [Google Scholar]
- 33.Dalton J.P., Neill S.O., Stack C., Collins P., Walshe A., Sekiya M., Doyle S., Mulcahy G., Hoyle D., Khaznadji E., et al. Fasciola hepatica cathepsin L-like proteases: Biology, function, and potential in the development of first generation liver fluke vaccines. Int. J. Parasitol. 2003;33:1173–1181. doi: 10.1016/S0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 34.Corvo I., Cancela M., Cappetta M., Pi-Denis N., Tort J.F., Roche L. The major cathepsin L secreted by the invasive juvenile Fasciola hepatica prefers proline in the S2 subsite and can cleave collagen. Mol. Biochem. Parasitol. 2009;167:41. doi: 10.1016/j.molbiopara.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Wang G.F., Peng D.L., Sun J.H., Huang W.K., Peng H., Long H.B. Cloning and sequence analysis of a new cathepsin L-like cysteine proteinase gene from Ditylenchus destructor. Chin. J. Biotechnol. 2011;27:60–68. doi: 10.13345/j.cjb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y.Y., Huang L., Wu X.Q. Bioinformatic and expression analysis of a cathepsin gene Bmcath1 in Bursaphelechus mucronatus. J. Nanjing For. Univ. 2015;39:12–16. doi: 10.3969/j.issn.1000-2006.2015.06.003. [DOI] [Google Scholar]
- 37.Hashmi S., Britton C., Liu J., Guiliano D.B., Oksov Y., Lustigman S. Cathepsin L is essential for embryogenesis and development of Caenorhabditis elegans. J. Biol. Chem. 2002;277:3477. doi: 10.1074/jbc.M106117200. [DOI] [PubMed] [Google Scholar]
- 38.Deng L.N., Wu X.Q., Ye J.R., Xue Q. Identification of autophagy in the pine wood nematode Bursaphelenchus xylophilus and the molecular characterization and functional analysis of two novel autophagy-related genes, BxATG1 and BxATG8. Int. J. Mol. Sci. 2016;17:279. doi: 10.3390/ijms17030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhoads M.L., Fetterer R.H. Developmentally regulated secretion of cathepsin L-like cysteine proteases by Haemonchus contortus. J. Parasitol. 1995;81:505–512. doi: 10.2307/3283844. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X.W., Wu X.Q., Huang L., Tian M.Q., Ye J.R. Specifically expressed genes of the nematode Bursaphelenchus xylophilus involved with early interactions with pine trees. PLoS ONE. 2013;8:e78063. doi: 10.1371/journal.pone.0078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang J.S., Lee H., Moon I.S., Lee Y., Koh Y.H., Je Y.H., Lim K.J., Lee S.H. Construction and characterization of subtractive stage-specific expressed sequence tag (EST) libraries of the pinewood nematode Bursaphelenchus xylophilus. Genomics. 2009;94:70–77. doi: 10.1016/j.ygeno.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 43.Rosso M.N., Dubrana M.P., Cimbolini N., Jaubert S., Abad P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe Interact. 2005;18:615–620. doi: 10.1094/MPMI-18-0615. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Xie H., Xu C.L., Li D.L., Zhang C. RNAi effect of cathepsin B gene on reproduction of Radopholus similis. Sci. Agric. Sin. 2010;43:1608–1616. doi: 10.3864/j.issn.0578-1752.2010.08.009. [DOI] [Google Scholar]
- 45.Li X.D., Zhuo K., Luo M., Sun L., Liao J. Molecular cloning and characterization of a calreticulin cDNA from the pinewood nematode Bursaphelenchus xylophilus. Exp. Parasitol. 2011;128:121–126. doi: 10.1016/j.exppara.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Wang X.R., Cheng X., Li Y.D., Zhang J.A., Zhang Z.F., Wu H.R. Cloning arginine kinase gene and its RNAi in Bursaphelenchus xylophilus causing pine wilt disease. Eur. J. Plant Pathol. 2012;134:521–532. doi: 10.1007/s10658-012-0035-0. [DOI] [Google Scholar]
- 47.Cardoso J.M.S., Fonseca L., Gomes P., Egas C., Abrantes I. Molecular characterization and functional analysis of a calponin gene from the pinewood nematode. Forest Pathol. 2015;45:467–473. doi: 10.1111/efp.12196. [DOI] [Google Scholar]
- 48.Koiwa H., Shade R.E., Zhu-Salzman K., D’Urzo M.P., Murdock L.L., Bressan R.A., Hasegawa P.M. A plant defensive cystatin (soya cystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera) FEBS Lett. 2000;471:67–70. doi: 10.1016/S0014-5793(00)01368-5. [DOI] [PubMed] [Google Scholar]
- 49.Lustigman S., Zhang J., Liu J., Oksov Y., Hashmi S. RNA interference targeting cathepsin L and Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Mol. Biochem. Parasitol. 2004;138:165–170. doi: 10.1016/j.molbiopara.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Shinya R., Takeuchi Y., Futai K. A technique for separating the developmental stages of the propagative form of the pine wood nematode, Bursaphelenchus xylophilus. Nematology. 2009;11:305–307. doi: 10.1163/156854108X399164. [DOI] [Google Scholar]
- 51.Ding X.L., Ye J.R., Wu X.Q., Huang L., Zhu L.H., Lin S.X. Deep sequencing analyses of pine wood nematode Bursaphelenchus xylophilus microRNAs reveal distinct miRNA expression patterns during the pathological process of pine wilt disease. Gene. 2015;555:346–356. doi: 10.1016/j.gene.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 52.He L.X., Wu X.Q., Xue Q., Qiu X.W. Effects of endobacterium (Stenotrophomonas maltophilia) on pathogenesis-related gene expression of pine wood nematode (Bursaphelenchus xylophilus) and pine wilt disease. Int. J. Mol. Sci. 2016;17:778. doi: 10.3390/ijms17060778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Regina W., Corinna W., Alexandra F., Jarutat T., Astrid H., Tobias B., William D., Barbara R. A Method for High Quality Digoxigenin-Labeled RNA Probes for in Situ Hybridization. [(accessed on 20 June 2018)]; Available online: http://www.ebiotrade.com/custom/upload/140506/1.pdf.
- 55.De Boer J.M., Yan Y., Smant G., Davis E.L., Baum T.J. In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 1998;30:309–312. [PMC free article] [PubMed] [Google Scholar]
- 56.Yu L.Z., Wu X.Q., Ye J.R., Zhang S.N., Wang C. NOS-like-mediated nitric oxide is involved in Pinus thunbergii response to the invasion of Bursaphelenchus xylophilus. Plant Cell Rep. 2012;31:1813–1821. doi: 10.1007/s00299-012-1294-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.