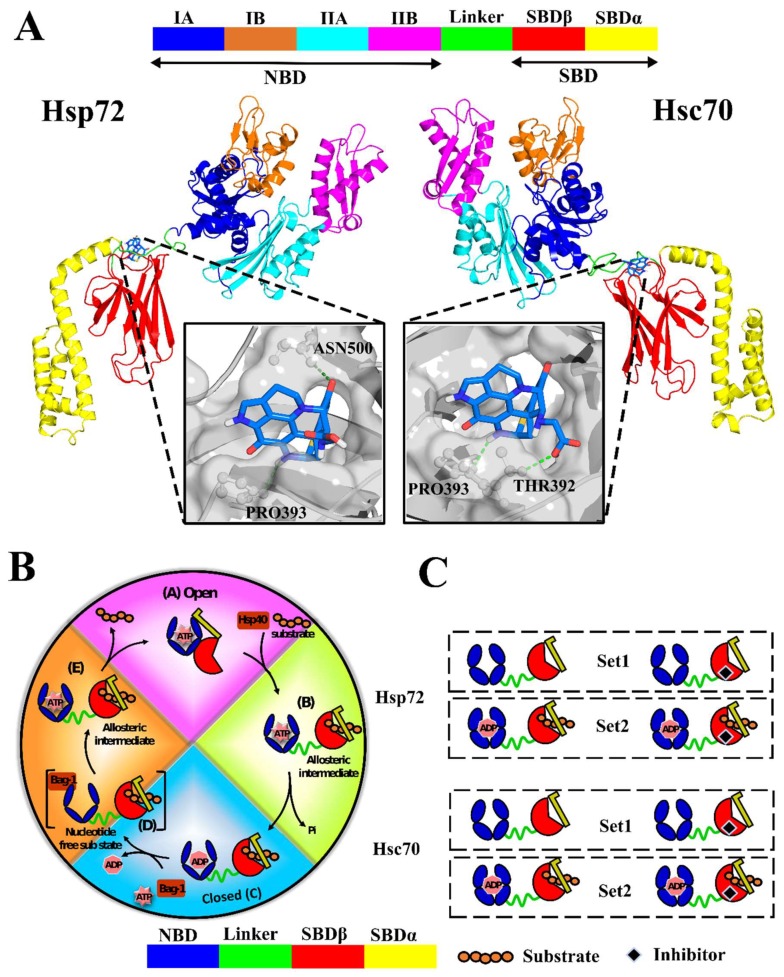
Figure 1.
(A) Cartoon representation of Hsp72 and Hsc70 3D structures calculated via homology modelling. The nearly full-length closed confirmation structures of Hsp72 and Hsc70 were modelled from Escherichia coli DnaK PDB entry: 2KHO. Predicted binding mode of SANC00132 docked to the allosteric target site, the SBDβ back pocket, is shown in sticks. Polar contacts are displayed as green dashes while interacting residues are shown in ball and stick. (B) Canonical nucleotide dependent cycle of Hsp70. (C) Schematic illustration of the experimental approach. Set1 comprises the simulation of SANC00132-free and SANC00132-bound systems only. Set2 simulations are for SANC00132-free and SANC00132-bound systems complexed with endogenous modulators (ADP and peptide substrate). Concisely, each protein was subjected to both sets of experiments.