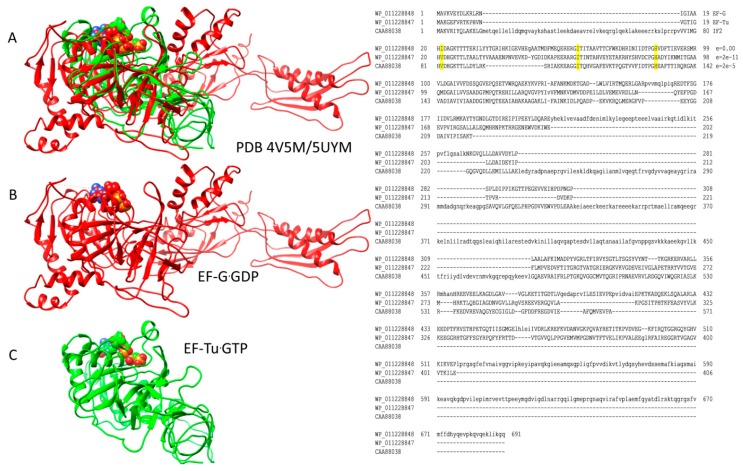
Figure 4.
EF-Tu and EF-G are homologs that occupy the same site on the ribosome; (A) overlay of ribosome-EF-Tu·GTP (green; PDB 5UYM) and ribosome-EF-G·GDP (red; PDB 4V5M). Overlays were done for ribosomal protein S2. Ribosomes are omitted from the images for simplicity; (B) EF-G·GDP structure (red); (C) EF-Tu·GTP structure (green). At the right, an alignment of Thermus thermophilus EF-G, EF-Tu, and IF2 is shown. Conserved residues (i.e., EF-Tu, Val21, Ile61, and His86) involved in stimulating GTP hydrolysis are indicated. In the alignment, e-values are versus Escherichia coli EF-G.