Abstract
γ-Amino butyric acid (GABA) mediated signaling is critical in the central and enteric nervous systems, pancreas, lungs, and other tissues. It is associated with many neurological disorders and craniofacial development. Glutamic acid decarboxylase (GAD) synthesizes GABA from glutamate, and knockdown of the gad1 gene results in craniofacial defects that are lethal in zebrafish. To bypass this and enable observation of the neurological defects resulting from knocking down gad1 expression, a photoactivatable morpholino oligonucleotide (MO) against gad1 was prepared by cyclization with a photocleavable linker rendering the MO inactive. The cyclized MO was stable in the dark and toward degradative enzymes and was completely linearized upon brief exposure to 405 nm light. In the course of investigating the function of the ccMOs in zebrafish, we discovered that zebrafish possess paralogous gad1 genes, gad1a and gad1b. A gad1b MO injected at the 1–4 cell stage caused severe morphological defects in head development, which could be bypassed, enabling the fish to develop normally, if the fish were injected with a photoactivatable, cyclized gad1b MO and grown in the dark. At 1 day post fertilization (dpf), light activation of the gad1b MO followed by observation at 3 and 7 dpf led to increased and abnormal electrophysiological brain activity compared to wild type animals. The photocleavable linker can be used to cyclize and inactivate any MO, and represents a general strategy to parse the function of developmentally important genes in a spatiotemporal manner.
Keywords: Glutamic acid decarboxylase, γ-amino butyric acid, GABAergic signaling, photoactivated morpholino, (8-bromo-7-hydroxyquinolin-2-yl)methyl, (8-cyano-7-hydroxyquinolin-2-yl)methyl
Introduction
γ-Amino butyric acid (GABA) mediated signaling is important in a number of neural and non-neural tissues in the vertebrate. Best known as an inhibitory neurotransmitter in the central nervous system (CNS), disruption in GABAergic signaling is associated with several neurological disorders, including epilepsy,1−3 schizophrenia,4−7 autism,8 spastic quadriplegic cerebral palsy 1,9 and stiff-person syndrome.10 Outside of the CNS, GABA and GABAergic signaling have been reported in the enteric nervous system11−14 and for tissues including the pancreas15,16 and lungs,17 among others.18 GABAergic signaling in the lungs plays a critical role in asthma,17 and disruption of GABAergic signaling in the pancreas appears to be causal for insulin-dependent diabetes mellitus.19−21
GABAergic signaling has also been implicated in craniofacial development in mammals. GABA is synthesized from glutamic acid by a reaction catalyzed by the enzyme glutamic acid decarboxylase (GAD, IUBMB Enzyme Nomenclature EC 4.1.1.15).22 In mammals, the majority of GAD protein exists as two molecularly distinct forms with molecular weights of 67 and 65 kDa, known as GAD67 and GAD65, which are encoded by the GAD1 and GAD2 genes, respectively.23,24 Whereas both enzymes make GABA from glutamate, they are found in different subcellular locations and at different expression levels in the same tissue, suggesting that each enzyme contributes differently to cell and neuron function.25−31GAD genes are also expressed in juveniles and adults, which indicates important roles in neuronal function and developmental processes.25,32−34 Because GAD converts an excitatory neurotransmitter into an inhibitory one, its function (or mis-function) has tremendous implications in the regulation of the nervous system. During development in mice and humans, loss-of-function mutations in the GAD1 gene, but not GAD2, or in the GABAA receptor, beta 3 (GABRB3) subunit have been reported to result in oral clefts (e.g., cleft lip and cleft palate), indicating that GABAergic signaling through GABAA is required for normal palate development in mammals.35−40
We are studying the role of GABAergic signaling in the developing zebrafish, with emphasis on the CNS. Zebrafish are an important and widely used model system for studying normal and abnormal physiology of vertebrates, especially embryonic development. Control of gene expression in the zebrafish is an invaluable tool for studying physiological processes.41−43 Genes important to development often have multiple functions at different life cycle stages. Mutations or techniques that interfere with early gene function often preclude the study of later functions. Further, irreversible changes to the genome made through knockout or transgenic organisms can result in compensation or embryonic lethality. An efficient method for manipulating gene expression in a spatiotemporal fashion would be a useful tool for investigating stage or tissue-specific roles of developmental genes.
In zebrafish, gene function can be disrupted by injecting embryos prior to the 16-cell stage with synthetic oligonucleotides called morpholino oligonucleotides (MOs),44 which contain DNA bases linked through a backbone consisting of phosphorodiamidates and morpholine. The difference in structure renders the MO resistant to nucleases, while still being able to hybridize to complementary mRNA sequences with high affinity and specificity. Depending on whether the MO binds to intron/exon sequences or near the start codon, RNA splicing or translation is blocked. The resulting gene silencing can last up to 4 days.44 MOs are incredibly important for genetic studies in zebrafish.45
Standard MOs induce global and immediate gene silencing in zebrafish, which limits their experimental utility. A photoactivatable MO provides control over the timing and location of gene silencing, enabling the study of the role of a gene’s expression at different stages of embryonic development. Photoactivatable negatively charged peptide nucleic acids (ncPNAs) and oligonucleotides have also been used to knock down gene expression in zebrafish,46−48 but MOs are more widely used in this important model organism. The first strategy reported for regulating the action of MOs in zebrafish with light used a hairpin linker49 that hybridized an inhibitor strand to the MO to inactivate it.50,51 Photolysis of the linker enabled dissociation of the MO from the inhibitor, releasing it to find its mRNA target. If the photocleaving element is (8-bromo-7-hydroxyquinolin-2-yl)methyl (BHQ),52,53 then two-photon excitation (2PE) activates the MO.51 PhotoMorphs use RNA as the inhibitor strand, which is photochemically cleaved into two pieces that bind poorly to the MO strand.54 Placing the photocleavage site in the middle of an MO strand enables destruction of the MO with light, leading to activation of gene expression at different time points in development.55 The bases on an MO strand can be directly photocaged with photoremovable protecting groups (PPGs), which blocks hybridization with the target mRNA, although the technique requires at least four PPGs to be distributed on the MO strand to prevent premature binding of the MO to the mRNA.56
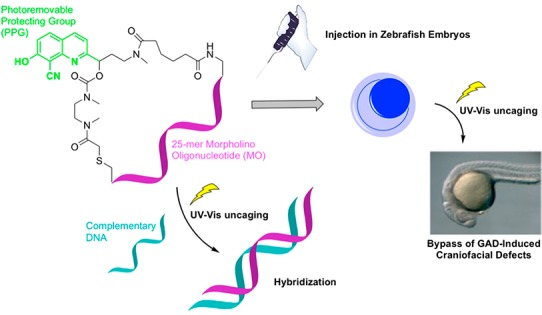
To eliminate the need for an inhibitor strand and ease of synthesis, Chen et al. created a caged cyclic morpholino (ccMO) against no tail-a (ntla).57 Cyclization of the MO through a photocleavable linker to generate a ccMO prevents the MO from binding to its mRNA target (Figure 1). A dimethoxynitrobenzyl (DMNB) PPG mediates the photocleavage of the linker. Tang et al. reported a ccMO against β-catenin-2 and ntl using a linker comprised of a nitrophenethyl (NPE) PPG.58 Subsequently, Chen and Deiters reported wavelength selective activation of ccMOs against ntla, spade tail (spt), and floating head (flh) using nitrobenzyl (NB) and diethylaminocoumarin (DEACM) based linkers for short (365 nm) and long-wavelength (470 nm) MO activation, respectively.59 Dmochowski et al. reported a ruthenium-based photolinker, RuBEP, for ccMOs against chordin (chd) and ntl that can be activated at 450 nm.106
Figure 1.

Photoactivation of a caged, cyclic morpholino (ccMO).
We prepared ccMOs against the gad1 and gad2 genes using a variation on Chen’s linker design that employs our quinoline-based PPGs BHQ and (8-cyano-7-hydroxyquinolin-2-yl)methyl (CyHQ),52,53,60 and demonstrated their use in conditionally manipulating gad gene function in the developing zebrafish embryo (see Methods for genetic nomenclature used in this report). Using BHQ and CyHQ expands the number of PPGs that can be used to prepare ccMOs activated with rapid photolysis kinetics (<1 μs time scale) using low intensity light to minimize phototoxicity. Further, BHQ and CyHQ can be photolyzed through two-photon excitation (2PE), which would enable ccMO activation using deeply penetrating IR wavelengths. In the course of investigating the function of the ccMOs in zebrafish, we discovered that zebrafish possess paralogous gad1 genes (VanLeuven, A. J.; Ball, R. E.; Gunderson, C. E.; Lauderdale, J. D., submitted). Other fish species have two gad1 genes, designated as gad1a and gad1b, and a single gad2 gene.61−63 In zebrafish, gad1a is located on chromosome 9, gad1b on chromosome 6, and gad2 on chromosome 24 (VanLeuven, A. J.; Ball, R. E.; Gunderson, C. E.; Lauderdale, J. D., submitted). Our gad MOs are designed to bind to the translation start sites of their respective transcript units. The gad1 MO is directed against gad1b, which is broadly expressed in the developing zebrafish CNS.25,64−66
Results and Discussion
Preparation of ccMOs
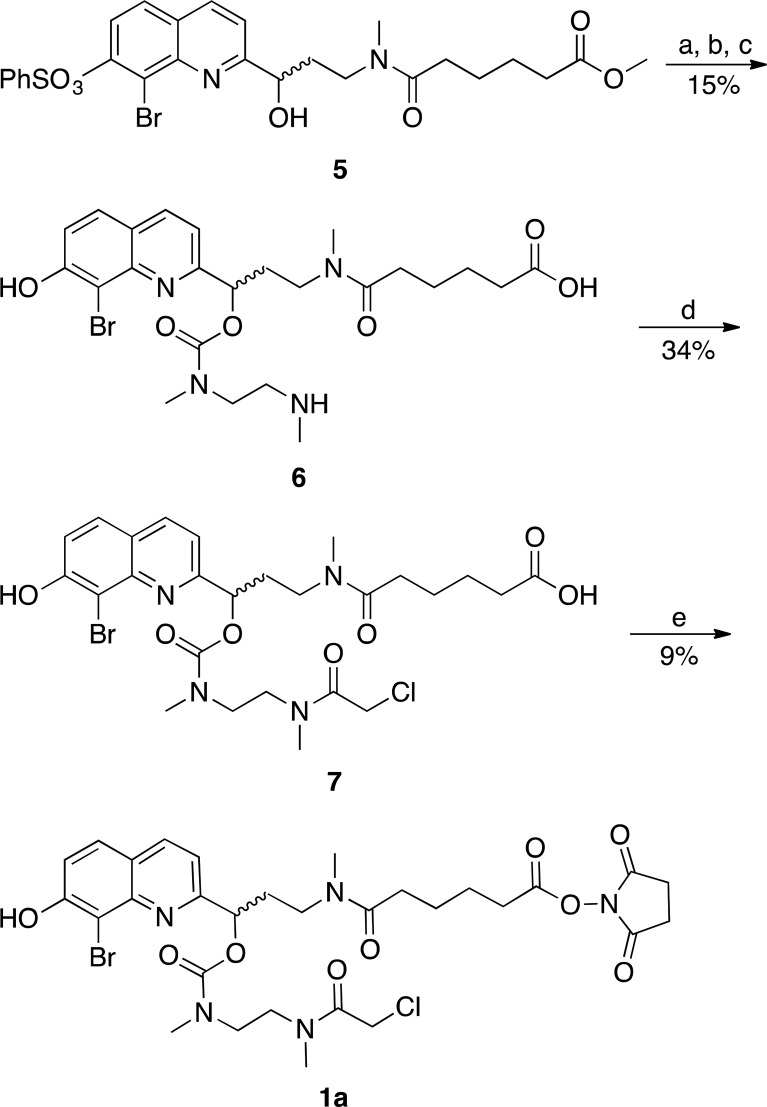
Two versions of the photocleavable linker were prepared: one from the BHQ PPG and the other from CyHQ. The BHQ-based linker 1a was prepared from the previously reported benzenesulfonate-protected BHQ derivative 5 (Scheme 1).51 The hydroxyl group in 5 was activated with carbonyl diimidazole and subsequently treated with N,N′-dimethylethylene-diamine to extend the chain. Treatment with base removed the benzenesulfonate and hydrolyzed the methyl ester to give the acid 6. The free secondary amine of 6 was acylated with chloroacetyl chloride to provide 7, whose carboxylic acid could be activated for coupling to the MO with disuccinimidyl carbonate, providing 1a.
Scheme 1. Synthesis of BHQ Linker 1aa.
Reagents and conditions: (a) carbonyl diimidazole, CH2Cl2, 15 min; (b) N,N′-dimethylethylene-diamine, CH2Cl2, 0 °C, 15 min; (c) acetonitrile/0.4 N NaOH(aq) (1:1), 12 h; (d) chloroacetyl chloride, DIEA, 15 min, then TFA, H2O, 12 h; (e) disuccinimidyl carbonate, pyridine, CH3CN, 12 h.
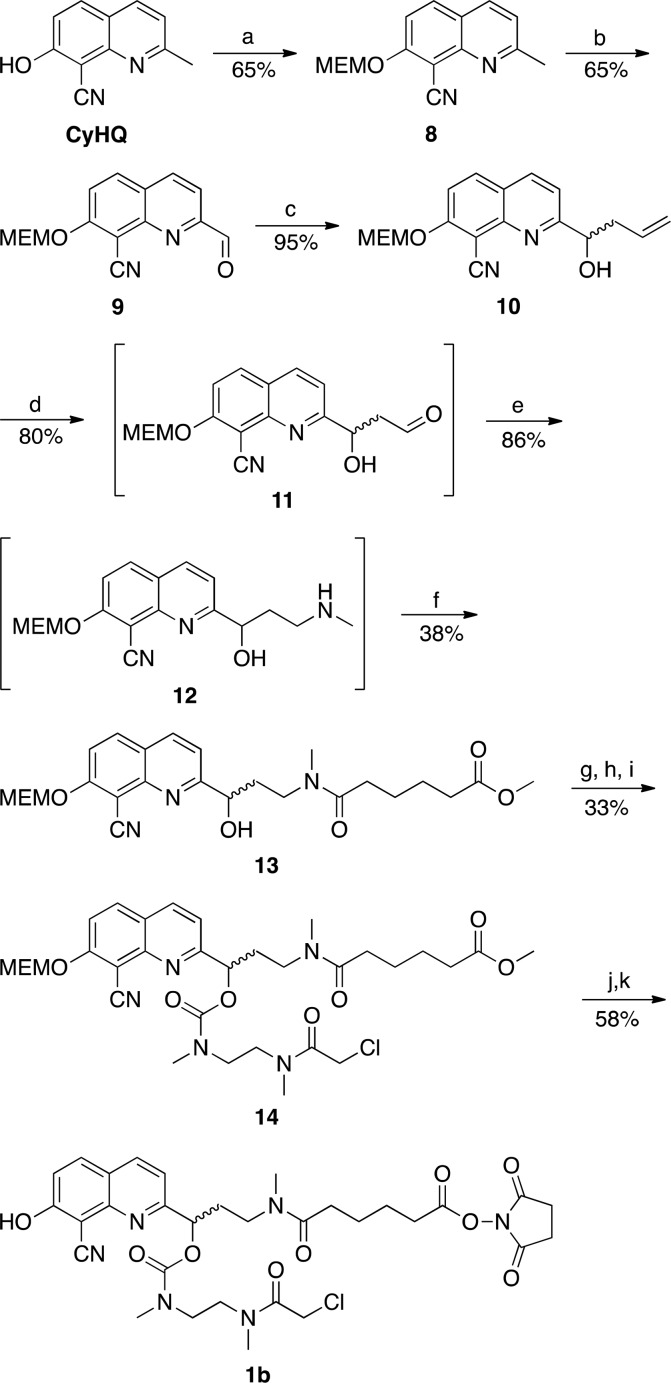
Linker 1b was prepared from CyHQ60,67 by first protecting the phenol with a MEM group to give 8, which was oxidized with selenium dioxide to aldehyde 9 (Scheme 2). An indium-mediated Barbier reaction with allyl bromide produced homoallylic alcohol 10, whose alkene could be oxidatively cleaved with osmium tetroxide to aldehyde 11. This aldehyde was too unstable in air to be isolated so it was immediately reductively aminated to 12, which was not purified, but quickly reacted with methyl adipoyl chloride to afford methyl ester 13. A similar method as described for 1a was used to convert 13 into 1b. A three-step sequence to grow a second arm of the linker from alcohol 13 provided 14. Trifluoroacetic acid removed the MEM group from the phenol and hydrolyzed the methyl ester, which could be converted to the succinimidyl ester 1b for coupling to a MO.
Scheme 2. Synthesis of CyHQ Linker 1ba.
Reagents and conditions: (a) MEMCl, iPr2NEt, CH2Cl2, 3 h; (b) SeO2, t-Bu-OOH, 1,4-dioxane, 45 °C, 3 h; (c) allyl bromide, indium, sat. NH4Cl(aq), THF, sonication, 55 °C, 90 min; (d) K2OsO4, 2,6-lutidine, NaIO4, 1:1 THF/H2O, 12 h; (e) methylamine (2.0 M in MeOH), AcOH, NaBH(OAc)3, THF, 12 h; (f) methyl adipoyl chloride, CH2Cl2, DIEA, 0 °C to rt, 6 h; (g) carbonyl diimidazole, CH2Cl2, 10 min; (h) N,N′-dimethylethylenediamine, CH2Cl2, 0 °C, 10 min; (i) chloroacetyl chloride, DIEA, 0 °C, 4 h; (j) trifluoroacetic acid/water (1:1), 40 °C, 12 h; (k) disuccinimidyl carbonate, pyridine, CH3CN, 12 h.
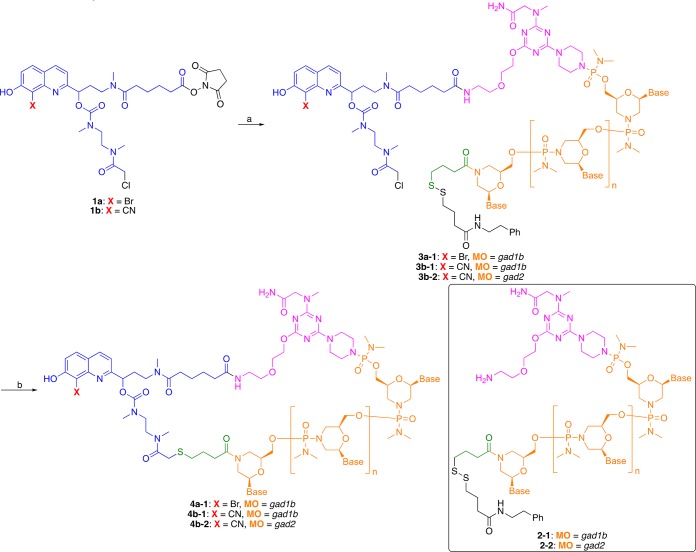
BHQ- and CyHQ-caged MOs against gad1b4a-1 and 4b-1 and gad24b-2, respectively, were prepared from their respective linkers 1a and 1b (Scheme 3). Linker 1a or 1b was coupled to the 5′-amine functionalized end of gad1b MO 2 through displacement of the succinimidyl ester in sodium borate buffer (pH 8.5). After desalting, 3a-1, 3b-1, or 3b-2 was cyclized by reducing the disulfide bond in situ with tris(2-carboxyethyl)phosphine (TCEP) to the sulfhydryl, which displaced the chloride on 3a-1, 3b-1, or 3b-2 to form the BHQ- or CyHQ-cMO (4a-1, 4b-1, or 4b-2) in ∼50% yield after purification by size exclusion chromatography (NAP-5 column eluting with a water mobile phase).
Scheme 3. Preparation of ccMOsa.
Reagents and conditions: (a) 0.1 M NaB4O7, pH 8.5, DMSO, functionalized MO with 5′ amino and 3′ disulfide (2–1 or 2–2); (b) tris (2-carboxyethyl)phosphine (TCEP), resin, 0.1 M Tris-HCl buffer, pH 8.4.
Photochemical Activation of gad1b MO
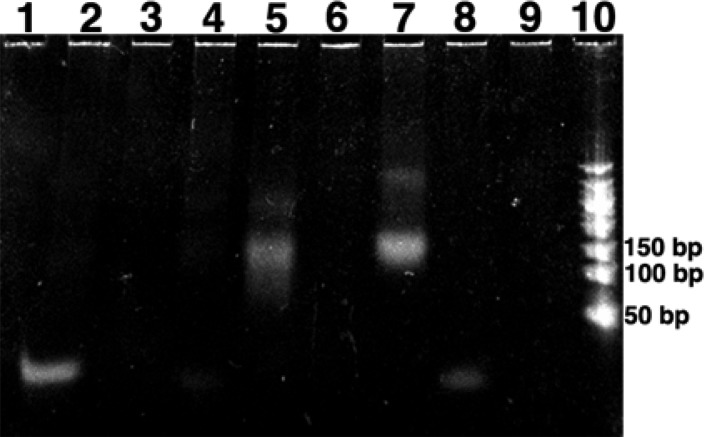
Photolysis of the ccMOs at 365 or 405 nm cleaved the linker, linearizing the cMO (Scheme 4). Gel shift assays were performed to determine the affinity of the gad1b CyHQ-protected cMO 4b-1 to its sequence specific target both before and after photolysis (Figure 2). Migration of the DNA and the DNA with the caged MO show bands commensurate with the 25 bp size of the fragment. The photolyzed ccMO and the control DNA bound to the underivatized MO show bands at the 150 bp range according to the ladder, because the oligomers are approximately twice the mass and possess half the charge. The result demonstrates that light exposure linearizes the gad1b MO, thereby facilitating its binding to the complementary DNA, which served as a surrogate for the gad1 mRNA.
Scheme 4. Photolysis of ccMOs.
Figure 2.
Photoactivation of the gad1b MO from CyHQ-gad1b-ccMO 4b-1. Results of gel shift assay of MO hybridization to its complementary DNA: lane 1, DNA; lanes 2–4, blank; lane 5, DNA and gad1b MO; lane 6, blank; lane 7, DNA, CyHQ-gad1b-ccMO, and light; lane 8, DNA, CyHQ-gad1b-ccMO, no light; lane 9, blank; lane 10, 50 bp DNA ladder.
An LC-MS/MS experiment was used to quantify the time course of photolysis of CyHQ-GAD1-ccMO (4b-1) at 405 nm (Figure 3), showing that the gad1b MO is activated rapidly with a quantum efficiency of 0.36 (Table 1). Quantitative evaluation of the photochemistry of BHQ-gad1b-ccMO (4a-1) was not performed because of its low molar absorptivity in the 365–405 nm range of wavelengths. Experiments in zebrafish (vide infra) show qualitatively photoactivation of 4a-1. The photochemical properties of CyHQ-gad2-ccMO (4b-2) were not assessed because knocking down gad2 does not cause craniofacial defects,68 and the compound was not used in the zebrafish studies. The gad1b and gad2 MO sequences differ substantially, demonstrating that a different MO sequence can be coupled to the photocleaveable linker 1b and supporting the generality of the strategy for creating photactivatable MOs.
Figure 3.
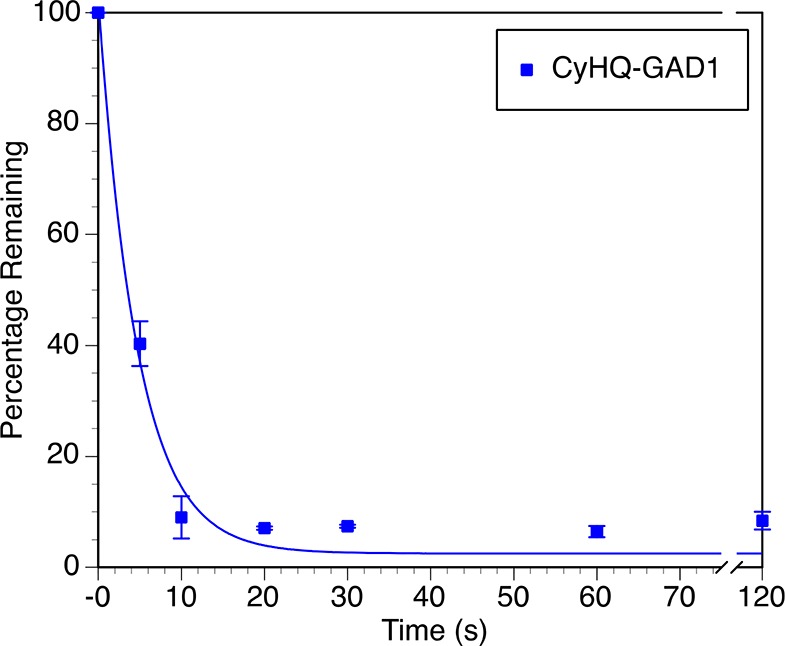
Time course of photolysis at 405 nm of CyHQ-gad1b-ccMO (4b-1) in water (50 μM). Each time point is the average of two experiments, and error bars represent the range of the measurement. Lines are least-squares fit of a simple decaying exponential. HPLC trace of CyHQ-gad1b-ccMO (4b-1) before and after photolysis is shown in Figure S3. Deconvoluted MS traces of CyHQ-gad1b-ccMO (4b-1) before and after light exposure are shown in Figures S4 and S5, respectively.
Table 1. Photophysical and Photochemical Data for ccMOs.
| λmax (nm) | ε (M–1 cm–1) | ε405 (M–1 cm–1) | t90% s) | I (einstein cm–2 s–1)a | Qua | sensitivity (εQu) | |
|---|---|---|---|---|---|---|---|
| 4a-1 | 265 | 26 420 | 460 | ||||
| 4b-1 | 365 | 26 580 | 3160 | 12.3 | 7.1 × 10–8 | 0.36 | 1214 |
| 4b-2 | 359 | 26 550 | 2650 |
Determined by potassium ferrioxalate actinometry.
Measured at 405 nm.
Stability of ccMOs in the Dark
CyHQ-gad1b-ccMO (4b-1) is relatively stable in the dark, having a half-life of 11.1 days in Tris buffer (pH 7.2) (Figure 4). Under red light and room light, the half-life decreases to 4.3 and 1.3 days, respectively. Because ccMOs need to work in a living organism, they will be exposed to degradative enzymes, so the stability of the ccMO, MO, and CyHQ-based linker 21 (Scheme S1) toward enzymatic degradation in the dark by representative degradative enzymes (proteinase K, Pronase E, esterase, and benzonase) was also evaluated using an LC-MS/MS assay (Figures S6–S8). CyHQ-gad2-ccMO (4b-2) was used instead of the gad1b version 4b-1, to conserve precious gad1b MO for biological studies. None of the enzymes tested caused degradation of CyHQ-gad2-ccMO (4b-2) more rapidly than that caused by the reaction buffers. Borate buffer (pH 8.0) was the most detrimental to 4b-2. The ccMO was generally more stable than the gad2 MO (2–2) and the pegylated linker 21. Given the similarity of the structures of 4b-2 and 4b-1, it is reasonable to assume similar stabilities.
Figure 4.

Time course of the degradation of CyHQ-gad1b-ccMO (4b-1) in Tris buffer (pH 7.2) under three different light conditions (dark, room light, and red light). Each time point is the average of two experiments and error bars represent the range of the measurement. Lines represent the least-squares fit of a single exponential decay.
CyHQ-gad1b-ccMO Enables Bypass of Craniofacial Defects
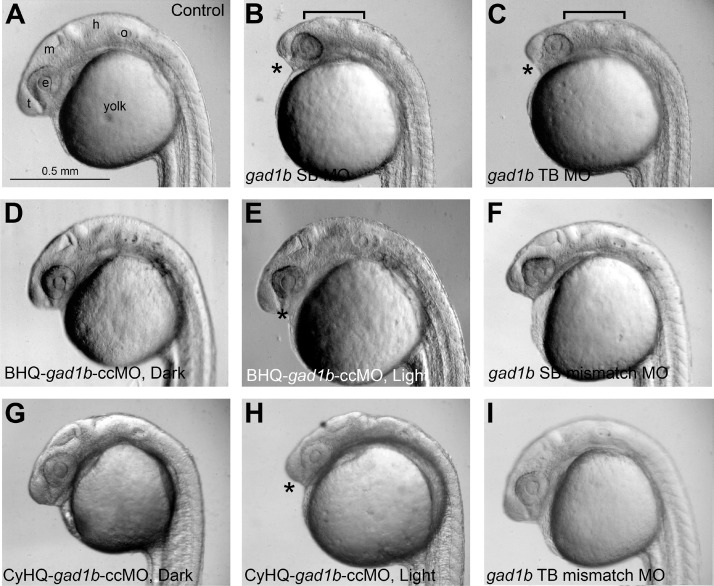
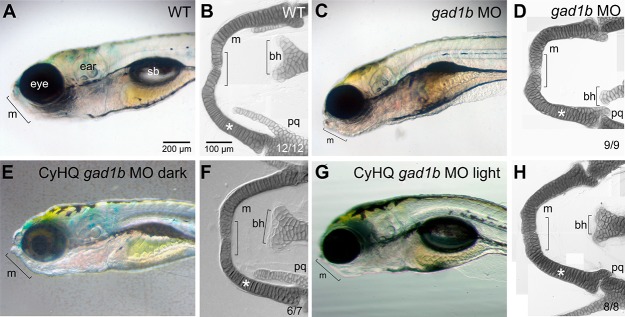
Zebrafish embryos injected at the 1–4 cell stage with gad1b MOs exhibited morphological defects in head development and outgrowth that were visible at 1 day post fertilization (dpf) (Figure 5; Figure S9). Changes visible by light microscopy in live animals included altered distribution of the presumptive head mesenchyme and altered gross structure of the midbrain-hindbrain region. By 3 days of development, the gad1b morphant embryos exhibited grossly normal brain structures, but exhibited aberrant jaw morphologies (Figure 6). Assessment of the cartilaginous head skeleton by Alcian blue staining in flat mount preparations at 7 dpf revealed defects in both the neurocranium and the pharyngeal skeleton in gad1b morphant larvae (Figure 7; Figures S10, S11, and S21). The neurocranium includes the ethmoid plate, trabeculae and parachordal cartilages, and the pharyngeal skeleton includes cartilages of the segmented jaw, opercular, and gill-bearing skeleton.23,24 Analysis of the neurocranium in gad1b morphants revealed midline defects of the ethmoid plate and shortened trabeculae (Figure S10). Analysis of the pharyngeal skeleton in gad1b morphants revealed midline defects in Meckel’s cartilage and the basihyal cartilage; these structures also exhibited altered cellular packing (Figure 7D,H; Figures S10–S12, S21).
Figure 5.
Knockdown of Gad1 causes defects in head development and outgrowth visible at 1 dpf (A) wild-type; (B) gad1b splice blocking MO; (C) gad1b translation blocking MO; (D,E) BHQ-gad1b-ccMO; (F) gad1b mismatch control for splice blocking MO; (G,H) CyHQ-gad1b-ccMO; (I) gad1b mismatch control for translation blocking MO. (D,G) BHQ- and CyHQ-gad1b-ccMO injected embryos raised in the dark exhibit morphological development comparable to wild-type embryos. (E, H) BHQ- and CyHQ-gad1b-ccMO injected embryos raised in the light exhibit morphological development comparable to embryos injected with gad1b splice blocking or translation blocking MOs (B–C). Asterisk denotes regions exhibiting altered distribution of the presumptive head mesenchyme; bracket denotes altered gross structure of the midbrain-hindbrain region. Normal development is observed in embryos injected with 5 base mismatch controls for the gad1b splice blocking MO (F) or translation blocking MO (I). Scale bar in (A) applies to all panels. t, telencephalon; e, eye; m, midbrain; h, hindbrain; o, otocyst.
Figure 6.
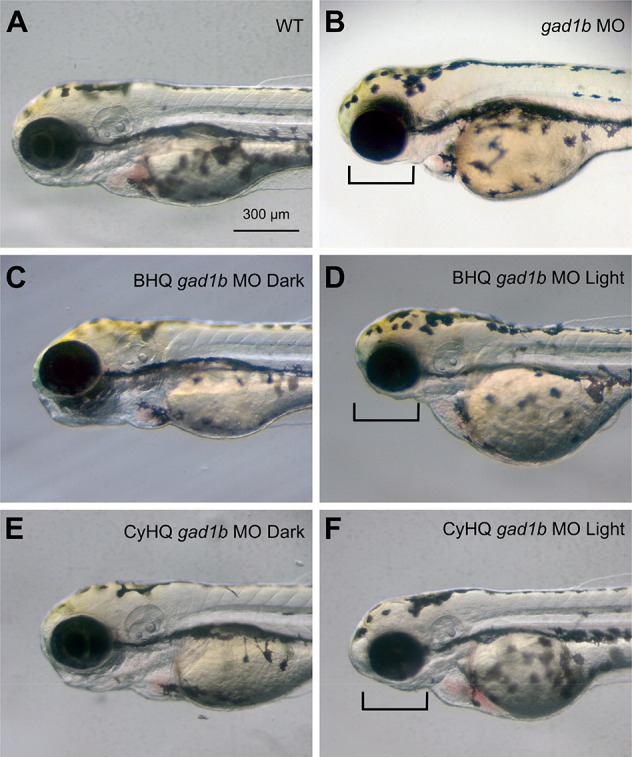
Knockdown of Gad1 causes jaw defects visible at 3 dpf (A) wild-type; (B) gad1b morphant; (C,D) BHQ-gad1b-ccMO; (E,F) CyHQ-gad1b-ccMO. (C,E) BHQ- and CyHQ-gad1b-ccMO injected embryos raised in the dark exhibit morphological development comparable to wild-type embryos. (B, D, F). BHQ- and CyHQ-gad1b-ccMO injected embryos raised in the light exhibit abnormal development of the presumptive lower jaw (region denoted by bracket) comparable to embryos injected with gad1b MOs; region includes Meckel’s cartilage. Scale bar in (A) applies to all panels.
Figure 7.
Morphological development of craniofacial features in the developing zebrafish probed by CyHQ-gad1b-ccMO. (A,B) Control fish. (B) Alcian blue stained, flat mount preparation showing normal development of Meckel’s (m), basihyal (bh), and palatoquadrate (pq) cartilages. (C,D) The gad1b morphant fish exhibit reduced Meckel’s and basihyal cartilages (denoted with brackets) with midlines defects. Note irregular organization of the cells in the Meckel’s cartilage of gad1b morphant fish (asterisk in D and H) compared to controls or ccMO injected fish photoactivated at 1 dpf.
The observed morphological defects were specific to gad1b MOs, and were observed using translation-blocking (Figures 5–7, S9–12, S21), splice-inhibiting (Figures 5 and S21), and photocaged gad1b MOs (Figures 5–7) that had been activated immediately after microinjection. Normal development was observed in embryos in which gad1b translation-blocking MOs were coinjected with synthetic gad1b mRNA immune to the MO (Figures S9–S11, S27) or injected with gad1b mismatch MOs (Figures 5 and S21). Embryos injected with BHQ-gad1b-ccMO (4a-1) or CyHQ-gad1b-ccMO (4b-1) under red-light conditions and grown in the dark exhibited grossly normal development (Figures 5 and 6) and exhibited normal expression of neural crest markers in premigratory and early migrating neural crest cells (Figure S24). Similarly, embryos injected with gad2 translation-blocking or splice-inhibiting MOs exhibited normal craniofacial development (Figure S9 and S12); however, gad2 morphants exhibit altered neurological activity (Figure S26). Western blot analyses revealed that, in the absence of synthetic mRNA, the MOs against gad1b and gad2 were effective in reducing below the limit of detection Gad proteins for the first 3–5 days of development (Figures S13–S20). These data suggest that the gad1b MO perturbs normal craniofacial development. Consistent with this, gad1b morphants exhibited altered expression of early neural crest markers in the head region of embryos just prior to the time of neural crest migration and in early migrating neural crest cells (Figure S22 and S24), and an altered distribution of presumptive migrating chondrogenic neural crest cells in embryos at 1 dpf (Figure S23). The gad1b morphants at 1 dpf also exhibited an increase in the number of apoptotic cells in the head and pharyngeal arch regions as visualized by acridine orange staining in live embryos (Figure S23).
The craniofacial phenotype in gad1b morphants is surprising because genetic knockouts of gad1b do not cause craniofacial defects (VanLeuven, A. J.; Ball, R. E.; Carpenter, J. M.; Filipov, N. M.; Lauderdale, J. D., submitted). Given our careful application of controls and rescue experiments following established guidelines to help distinguish specific phenotypes from off-target effects associated with MOs,69 we conclude that the gad1b MO-induced craniofacial phenotype is due to perturbation of GABAergic signaling and represents a more severe phenotype than in gad1b null mutants, as has been observed with other MO-induced phenotypes compared to the corresponding mutants.70 Consistent with this idea, treatment of early stage embryos with GABAA receptor antagonists alters expression of early neural crest markers in the head region of embryos in a manner similar to that observed in gad1b morphants. One explanation is that the gad1b MO is affecting expression of both gad1a and gad1b. Because they have similar mRNA sequences at the translation start site,61 the gad1b MO may knockdown expression of both gad1a and gad1b. If this is the case, we hypothesize that zebrafish embryos null for gad1a and gad1b will exhibit the craniofacially defective phenotype. Conversely, the gad1b translation blocking MO is an exact match for the sequence around the gad1b translation initiation ATG (Table 2). When compared to gad1a, the MO matches 19/25 nucleotides, which would be expected to significantly reduce the binding affinity with gad1a. Many studies make use of a five-base mismatch control MO. A recent study investigating off-target effects of MOs found that 11 consecutive bases complementary to a target sequence were sufficient for binding and subsequent blocking of splice sites.71 The gad1b translation blocking MO used in our study has only 8 consecutive bases complementary to gad1a, so it seems unlikely that the gad1b translation blocking MO used here would also block translation of gad1a. Nevetheless, this cannot be directly tested at this time, because an antibody specific to Gad1a has yet to be identified.
Table 2. Start Sequence Comparison between gad1a, gad1b, and the gad1b MOa.
| gad1b | 5′-GGACTGATGGCGTCTTCTGCACCTT-3′ | (25/25 match) |
| gad1b MO | 3′-CCTGACTACCGCAGAAGACGTGGAA-5′ | |
| gad1a | 5′-GaggTGATGGCGgCgTCTGCACCcT-3′ | (19/25 match) |
Mismatched nucleotides in gad1a compared to the gad1b MO are denoted in lower case. The start codons for gad1b and gad1a are denoted in bold.
Another explanation for the craniofacial phenotype is that the gad1b MO alters isoform expression in a developmentally inappropriate way, and it is this change that leads to altered cartilage development. We detected different isoforms of Gad1 protein in the developing zebrafish, and the pattern of isoform expression changed as a function of development (Figure S13). Interestingly, gad1b morphants exhibited a shift in gad1 isoform expression during the period when the MO no longer fully suppressed translation (Figure S16). Although unexpected, this latter mechanism may provide novel insight into gad gene function in non-neural tissues.
To test if we could use photocaged ccMOs to knockdown gad1b activity while bypassing craniofacial deformities, photoactivation was carried out at 1 day of development. This time point was chosen for two reasons. First, we hypothesized based on the head morphology of gad1b morphants at 1 dpf that the defects in the cartilages of the head skeleton were due to perturbation of the cranial neural crest (CNC) cells either before or at the time that these cells were migrating to populate the pharyngeal arches. Beginning at approximately 15 hpf, CNC cells delaminate from the ectoderm overlying the dorsal neural tube in a wave originating at the midbrain and progress along in caudal direction and migrate as separate streams into the pharyngeal arches.39,40 The bulk of this migration occurs between 18 and 24 hpf. Neurocranial precursors originate in the midbrain region and migrate between the eyes to form the palatal shelves, and the pharyngeal arches are populated by CNCs that emigrate from the hindbrain.65 Second, we wanted to knockdown gad1b activity in neurons in the developing brain and spinal cord. By 1 dpf, gad1b expressing neurons are detectable at all axial levels of the CNS.25,64−66
The hypothesis that gad1 is expressed in cranial neural crest cells in zebrafish . Mice homozygous for mutations in GAD1, but not GAD2, or in the GABRB3 subunit develop a cleft secondary palate.35−38 Administration of GABA to GAD67-deficient mouse fetuses resulted in restoration of the first steps of palate formation.72 In humans, mutations in GABRB3 and in the GAD1 comes from human and mouse datatranscript unit have been linked with nonsyndromic cleft lip with or without cleft palate.39,40 GABA produced by nonneuronal cells in the palatal epithelium mediates cell proliferation during palatal shelf elevation and subsequently cell migration and/or differentiation during palatal shelf fusion.73 During palatogenesis in mice, GAD and GABA are expressed in the palatal epithelium of the vertical shelves, in the medial edge epithelium of the horizontally oriented palatal shelves, and in the epithelial seam during fusion.73−75 Ubiquitous expression of the GABAA β3 subunit gene rescued the cleft palate in GABRB3 mutant mice,76 neuronal-specific transgene expression did not.73 In mice, too much GABA signaling77−79 or too little72 disrupts palate formation, and a link between benzodiazepine (a potentiator of GABA signaling) use during pregnancy and craniofacial defects has been reported in humans,80−82 but is controversial.83 These data suggest the process during palatogenesis requires a specific range of GABA signaling during a critical period for normal development; however, the underlying cellular and molecular mechanism are not known. Although secondary palate development does not appear to occur in fish,84,85 several lines of evidence are consistent with the idea that GABAergic signaling (and Gad1) is playing a role in the cranial neural crest. In addition to the knock-down data presented (vide supra), treatment of embryos with pentylenetetrazole (PTZ), a GABAA antagonist,86 phenocopies the increase in cells expressing neural crest markers in the brain comparable to that observed in the morphants (Figure S25). The gad1b gene is expressed in the early embryo as detected by RT-PCR (VanLeuven, A. J.; Ball, R. E.; Gunderson, C. E.; Lauderdale, J. D., submitted).
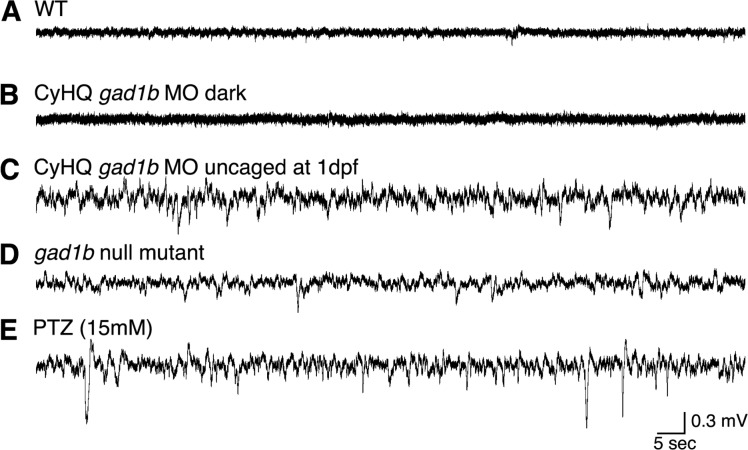
Zebrafish embryos were injected at the 1–4 cell stage with CyHQ-gad1b-ccMO (4b-1) under red-light conditions and raised in the dark for the first day of development. Half the injected embryos were then photoactivated and returned to the dark. All embryos were assessed for craniofacial development at 1, 3, and 7 dpf (Figure 7) and for behavioral and electrophysiological correlates of seizure activity at 3 dpf (Figure 8). Zebrafish harboring the gad1b cMO activated at 1 dpf exhibited normal or near-normal development of the cartilaginous head skeleton (Figure 7), behavioral correlates of seizure (data not shown), and increased and abnormal electrophysiological brain activity compared to wild type animals (Figure 8), consistent with a reduction in GAD function. The electrophysiological traces of the cMO morphants were comparable to those obtained from larvae null for gad1b and wild-type fish exposed to PTZ, which causes seizures in zebrafish.87,88 PTZ acts by blocking the GABAergic systems, in particular inhibition of GABAA receptor-mediated inhibitory postsynaptic potentials.86 It has been widely used as a kindling model of epilepsy in rodents89 that has recently been extended to frogs and fish.87,90 The differences in the voltage amplitude between the recording from the gad1b–/– fish (Figure 8D) and the CyHQ-GAD1-ccMO + light fish (Figure 8C) might reflect the more severe effect of the MO.
Figure 8.
Zebrafish gad1b morphants demonstrate abnormal electrophysiological brain activity. Baseline electrophysiological field recordings from the brain were made at 3 dpf on (A) wild-type zebrafish, (B) fish grown in the dark from CyHQ-GAD1-ccMO (4b-1) injected embryos, (C) fish grown from CyHQ-gad1b-ccMO (4b-1) injected embryos and exposed to light at 1 dpf, (D) fish grown from gad1b null mutant embryos (gad1b–/–), and (E) wild-type fish exposed to 15 mM pentylenetetrazole (PTZ).
Conclusions
Using a photocleavable linker, we prepared BHQ- and CyHQ-based ccMOs against gad1b and gad2. The most efficiently photolyzed compound, CyHQ-gad1b-ccMO (4b-1), had a quantum efficiency of 0.36 and was completely linearized after a brief exposure to 405 nm light, thereby activating it toward hybridization with its complementary gad1b mRNA partner. The ccMOs were sufficiently stable in the dark to enable studies in zebrafish that last several days. Importantly, they showed remarkable resistance to representative degradative enzymes proteinase K, Pronase E, esterase, and benzonase. The high pH buffers used in each of the assays were the sole cause of ccMO degradation; the enzymes had no degradative effects on the ccMOs. Zebrafish embryos injected with BHQ-gad1b-ccMO (4a-1) or CyHQ-gad1b-ccMO (4b-1) and grown in the dark developed normally, effectively bypassing the embryonically lethal effect of knocking down gad1 expression with a gad1b MO, and enabling the observation of the abnormal neurological effects of knocking down gad1b expression in older animals. The photocleavable linker can be cyclized with any MO, so this represents a general strategy for studying the role of embryonically lethal genes at later stages of development in zebrafish.
Methods
Genetic Nomenclature
Standard gene nomenclature is used. For zebrafish, gene symbols are lowercase and italicized and protein symbols are capitalized. For other vertebrates, human conventions are used where gene symbols are in all capitals and italicized and protein symbols are in all capitals. With respect to the full name of the ccMOs, the gene symbol is formatted to be consistent with that of the linker
Synthesis
See the Supporting Information for detailed experimental procedures to prepare BHQ linker 1a, CyHQ linker 1b, and CyHQ-linker-PEG 21.
BHQ-gad1b-MO (3a-1), CyHQ-gad1b-MO (3b-1), and CyHQ-gad2-MO (3b-2)
A 25-base gad1 or gad2 MO oligomer with 5′-amine and 3′-disulfide functionalization (2-1, 5′-AAGGTGCAGAAGACGCCATCAGTCC-3′, or 2-2, 5′-GAAACCAAAACCCGTGTGATGCCAT-3′; 50 or 100 nmol; Gene-Tools) was dissolved in 0.1 M Na2B4O7, pH 8.5 (100 μL). Photocleavable linker 1a or 1b (1.0 μmol, 67 equiv) in DMSO (15.0 μL) was added, and the reaction was shaken in the dark until LC-MS indicated that it was complete. The resulting mixture was purified on a NAP-5 gel-filtration column and lyophilized; the resulting white solid was dissolved in water (200 μL). Acetic acid (2.00 μL) was added to the solution, which was then washed successively with chloroform (3 × 200 μL) and EtOAc (2 × 200 μL), and then neutralized with 10% aq. NH4OH. The solution was lyophilized to dryness, affording the conjugated product 3a-1, 3b-1, or 3b-2 as a white solid, which was taken directly to the next step without further purification. BHQ-gad1b-MO (3a-1): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C349N165O106H534P25ClBrS2 9688, found 9688. CyHQ-gad1b-MO (3b-1): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C342N166O106H524P25ClS29531, found 9531. CyHQ-gad2-MO (3b-2): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C342N163O106H525P25ClS29490, found 9490.
BHQ-gad1b-ccMO (4a-1), CyHQ-gad1b-ccMO (4b-1), and CyHQ-gad2-ccMO (4b-2)
Gel-immobilized TCEP (100 μL) (Pierce Biotechnology) was washed with 0.1 M Tris-HCl buffer, pH 8.4 (3 × 100 μL) in a centrifuge filter tube. BHQ-GAD1MO (3a-1) or CyHQ-GAD1MO (3b-1) was dissolved in 0.1 M Tris-HCl buffer, pH 8.4 (100 μL) and added to the washed gel; the reaction was shaken for 10 h in the dark. The supernatant was collected by centrifuge at 1000 rpm for 30 s. The gel slurry was washed with 0.1 M Tris-HCl buffer, pH 8.4 (3 × 100 μL), and the eluted fractions were combined with the supernatant. This mixture was purified on a NAP-5 gel-filtration column and lyophilized to afford 4a-1 (8.2 nmol, 17% over 2 steps), 4b-1 (35.1 nmol, 35% over 2 steps), or 4b-2 (29.0 nmol, 29% over 2 steps). BHQ-gad1b-ccMO (4a-1): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C337N161O105H518P25BrS 9431,found 9433. CyHQ-gad1b-ccMO (4b-1): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C338N165O105H516P25S 9377, found 9376. CyHQ-gad2-ccMO (4b-2): MS-ESI-LC-MS-Q-TOF (m/z) [M]+ calcd for C338N162O105H517P25S 9336, found 9336.
Photochemistry
Gel Shift Assay
Solutions (0.88 μM) of nonderivatized MO, its antisense DNA (5′-TTCCACGTCTTCTGCGGTAGTCAGG-3′), irradiated ccMO (10 min exposure to light from a mercury lamp to achieve complete photolysis), and nonirradiated ccMO were prepared in TBE buffer. Light-sensitive materials were handled in the dark or under red safety lamps. Four samples were prepared as follows: DNA solution (8 μL), DNA solution (8 μL) and MO (8 μL), DNA solution (8 μL) and photolyzed ccMO (8 μL), and unphotolyzed ccMO (8 μL) and DNA solution (8 μL). Hybridization was performed by heating the samples in a water bath (45 °C) for 30 min followed by cooling on ice. Loading buffer (3 μL, 5×) was added to each sample, which was subsequently loaded onto 12% native PAGE gel. The gel was run with a 50 bp ladder (GelPilot, Qiagen) at 40 V for 3.5 h. Gels were stained with SYBR Gold nucleic acid gel stain (Thermo Fisher Scientific) for 30 min for visualization.
Quantum Efficiency
Compound 1b was dissolved in water, and an aqueous solution of gad1b MO (internal standard) was added to a final concentration of 50 μM for both analytes. Aliquots (50 μL) of the solution were placed in a microcuvette (26.10F-Q-10, Starna, Atascadero, CA, 10 × 1 × 1 mm3 illuminated dimensions) and irradiated for different time intervals (0, 5, 10, 20, 30, and 60 s) with a LED lamp (Cairn OptoLED Lite) at 405 nm. Each time point was repeated in duplicate. The irradiated solutions were analyzed by LC-MS/MS (Agilent UHD Accurate-Mass QTOF LC/MS system 6540, Agilent Technologies, Palo Alto, CA) to quantify the residual concentration of 1b. Chromatographic profiles were obtained using an Agilent PLRP-S C18 column 150 × 4.6 mm with 8 μm particle size and gradient elution: eluent A being acetonitrile and eluent B consisting of water. The analysis started with 2% of eluent A, which was linearly increased up to 100% in 20 min, then remaining at 100% until 30 min. The flow rate was 0.5 mL min–1, and injection volume was 10 μL. Quantification of the eluted material was obtained by developing an appropriate MS-MS method using the QTOF detector. Collision induced dissociation was performed with a fixed collision energy set at 40 eV and two transitions were recorded at m/z 1563.8 and 1849.5, operating the MSD in positive ion mode. Quantitative data were obtained by comparison to standard calibration curves using the Agilent MassHunter Workstation Software for Quantitative Analysis. The percentages of compound left in solution were plotted versus time of irradiation and fitted to a single exponential decay curve using the software DeltaGraph (Red Rock Software, Inc.). The quantum efficiency (Qu) of the photolysis reaction was determined as previously described,53 using the following equation: Qu = (Iσt90%)−1,91 where I is the intensity of the lamp in einstein cm–2 s–1 (measured by ferrioxalate actinometry92), σ is the decadic extinction coefficient (1000ε, molar extinction coefficient), and t90% is the time in seconds required for 90% of the starting material to be photolyzed.
Stability Studies
Stability in the Dark, Red Light, and Room Light
CyHQ-gad1b-ccMO (4b-1) was dissolved in Tris buffer (150 mM KCl, 10 mM Tris-HCl, pH 7.2, phenol red 0.01%) to a final concentration of 100 μM. Aliquots of the solution (50 μL) were placed in microcentrifuge tubes and exposed to different light conditions (dark, room white light, red light). The tubes were sampled (6 μL) after different time intervals and aqueous solutions of the internal standard (gad1b MO100 μM) were added (6 μL) to a final concentration of 50 μM for both analytes. Quantitative data were obtained with the same LC-MS/MS method described for the photolysis experiment. The experiment was performed in duplicate. The percentage of CyHQ-gad1b-ccMO (4b-1) remaining in solution was plotted against time and fitted to a single exponential decay curve with the software DeltaGraph (Red Rock Software Inc.). Three different curves were generated, representing the three conditions evaluated (dark, room white light, red light). Half-life (t1/2) values were obtained from the corresponding curve equation.
Stability Toward Enzymatic Degradation
CyHQ-gad1b-ccMO (4b-2) (3 nmol) was dissolved in the appropriate buffer (Table 3) and a fresh solution of enzyme in buffer was added. Aliquots of the resulting solution (50 μL) were placed in microcentrifuge tubes and incubated at the appropriate temperature (Table 3) in the dark. A control experiment without enzyme was also performed under the same conditions. The tubes were sampled (10 μL) after different time intervals (0, 2, and 4 days) and combined with aqueous solutions of the internal standard (gad2 MO) (10 μL). Quantitative data were obtained with the same LC-MS/MS method described for the photolysis experiments. Each experiment was performed in triplicate. The same experimental conditions were also used to evaluate the enzymatic stability of the two separate portions of CyHQ-gad1b-ccMO (4b-2): the ad-hoc synthesized linker portion (CyHQ-linker-PEG, 21) and the MO alone (gad2 MO, 2-2).
Table 3. Experimental Conditions for the Enzymatic Stability Experiment.
| enzyme | buffer | enzyme concen | temperature |
|---|---|---|---|
| proteinase K | Tris 50 mM, CaCl2 5 mM, pH 7.8 | 0.4 mg/mL (0.6 U/reaction) | 37 °C |
| pronase E | Tris 50 mM, CaCl2 5 mM, pH 7.8 | 1.4 mg/mL (0.2 U/reaction) | 37 °C |
| esterase | borate 10 mM, pH 8.0 | 3.3 mg/mL (2.5 U/reaction) | 25 °C |
| benzonase | Tris 20 mM, MgCl2 2 mM, NaCl 20 mM, pH 8.0 | 0.5 μL (125 U/reaction) | 37 °C |
A positive control experiment was run in parallel for each enzyme to ensure that the hydrolase was active under the assay conditions. The control substrate for the proteolytic and esterolytic enzymes was N-acetyl-l-tyrosine ethyl ester (Ac-Tyr-OEt), bearing both the ester and amide functionality. Almost complete hydrolysis (>90%) of Ac-Tyr-OEt was observed after 10 min of incubation in all cases (HPLC). An in-house DNA sequence was used as a positive control for the benzonase stability experiment. Degradation of the DNA was monitored by UV–vis spectroscopy (ΔA265) with a NanoDrop spectrophotometer. A 50% increase of absorptivity was found after 1 h of incubation with benzonase, confirming DNA hydrolysis.
Zebrafish Studies
Zebrafish Maintenance
Adult and larval zebrafish (Danio rerio) were obtained from lines maintained in the University of Georgia Zebrafish Facility following standard procedures.93 Embryos and larvae were staged using standard staging criteria.93,94 Wild-type fish of the WIK strain were originally obtained from the Zebrafish International Research Center (ZIRC). The line of zebrafish mutant for gad1b was generated by CRISPR-Cas9 mediated genome editing of the gad1b locus. Additional details will be provided in a separate publication (VanLeuven, A. J.; Ball, R. E.; Carpenter, J. M.; Filipov, N. M.; Lauderdale, J. D., submitted). The gad1bgav2303/gav2303 allele used in these studies harbored a 10 bp deletion in exon 4 and was a functional null mutation. All adult fish were maintained in an Aquatic Habitats (Apopka, FL) multirack system. Habitat water consisted of reverse osmosis filtered/sterilized water to which sodium bicarbonate and other salts (Instant Ocean, Aquarium Systems, Inc., Mentor, OH) were added to maintain pH from 7.0 to 7.4 and conductivity between 400 and 430 μS. All experimental procedures were conducted in accordance with National Institutes of Health guidelines for use of zebrafish in research under protocols approved and overseen by the University of Georgia Institutional Animal Care and Use Committee.
Morpholino Microinjection and mRNA Rescue
MOs were microinjected using pressure into one to four-cell stage embryos following standard protocols69,70,95,96 under red-light conditions. Approximately 1 nL was injected into each embryo at the following concentrations: gad1 MO, 0.3 ng/nL; gad2 MO 1 ng/nL; BHQ- or CyHQ-gad1-ccMO, 0.35–0.45 ng/nL. See Figures S14–S17 for titration data of gad1b and gad2 MOs. The following 5 base mismatch controls were used: gad1b translation blocking mismatch MO, 5′-AAGCTCCAGAACACCCCATCACTCC-3′; gad2 translation blocking mismatch MO 5′-GATAGCAAAAGCCGTCTCATGCCAT-3′. In rescue experiments, a synthetic gad1b mRNA was coinjected with the gad1b MO. The synthetic gad1b mRNA lacked the gad1b MO target sequence while preserving the amino acid sequence; specifically, the sequence was changed to 5′ GAATTCACCATG GCA AGC AGC GCT CCA TCT... where the nucleotides in bold were changed from the endogenous sequence and the translation initiation site is underlined. See Figure S22 for titration data of synthetic gad1 mRNA.
Drugs
The pentylenetetrazol (PTZ) used for this study was purchased from Sigma-Aldrich (cat. #P6500, St. Louis, MO). Stock PTZ solutions of 300 mM were prepared in distilled water following protocols used for administration to mammals97 and stored in aliquots at −20 °C. Compounds dissolved in embryo medium are readily absorbed through the gastrointestinal tract or across the skin of larvae.98
Electrophysiology
Electrophysiological recordings were collected from 3 dpf zebrafish following protocols established for 7 dpf larvae.87 Briefly, zebrafish larvae were immobilized by exposure to 250 μM α-bungarotoxin (Life Technologies/ThermoFisher, cat. #B1601)99 in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, pH 7.2)100 and mounted dorsal-side up in 1.2% agarose (Thermo Fisher Scientific, Waltham, MA) in E3 medium. After embedding, larvae were placed on the upright stage of a Zeiss Examiner.Z1 microscope (Carl Zeiss Microscopy, LLC One Zeiss Drive Thornwood, NY 10594) and bathed with fresh E3 media. Under direct visual guidance, a glass microelectrode (approximately 1 μm tip diameter, 5–10 MΩ impedance) was placed in the optic tectum. The optic tectum was chosen to facilitate comparison with previously published data obtained from larval zebrafish.87 Electrodes were were filled with 2 M NaCl. A chloride-coated silver wire (0.010 in., A-M Systems, Inc. Sequim, WA) reference electrode was placed touching the medulla or spinal cord. Field recordings were collected using an Axoclamp 900a amplifier (Axon Instruments, Union City, CA). The amplified voltage was passed through a Hum Bug Noise Eliminator (AutoMate Scientific, Berkley, CA), band-pass filtered from 1 to 0.1 kHz, and digitized at 10 kHz using a Digidata 1440 interface and stored on a PC using pClamp software (version 10.3, Molecular Devices). Experiments were carried out at 23 ± 0.5 °C. To chemically induce seizures, larvae immobilized in agar were exposed to E3 medium containing 15 mM PTZ, which is a concentration shown to reliably induce clonus-like seizure behavior.87
Western Blot Analysis
Western analysis was performed following standard protocols.101,102 To determine MO knockdown of gad gene encoded proteins, 30–100 embryos per injection set were collected at 3 dpf, deyolked in ice-cold calcium-free Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 5.0 mM HEPES, pH 7.2),93 and homogenized in a solution containing 60 100 μL RIPA buffer (cat. #89900, Thermo Fisher Scientific Inc., Waltham, MA) with 1 mM EDTA solution and 1 mM protease inhibitor cocktail (cat. #P8340, Sigma-Aldrich, St. Louis, MO). Samples were centrifuged for 10 min at 18000g and the supernatant collected and stored at −80 °C until further use. Protein concentration was measured by Bradford assay.103 Samples containing 10 μg of total protein were separated by SDS-PAGE and transferred to 0.45 μm nitrocellulose membrane (Bio Rad, Hercules, CA). Blots were probed using a mouse monoclonal to GAD67 (cat. #sc-58531, Santa Cruz Biotechnology Inc.) at a 1:200 dilution, a rabbit polyclonal to Gad2 (cat. #55772, AnaSpec, Inc., Freemont, CA) at a 1:1000 dilution and a rabbit polyclonal to GAPDH (ab9484, Abcam, Cambridge, MA) at a 1:10,000 dilution. Bands were visualized by chemiluminescence (Immun-Star WesternC chemiluminescence kit, Bio Rad, Hercules, CA).
Alcian Blue Staining of Cartilages
Cartilage staining was performed using a modified protocol of Kimmel et al.104 Briefly, larvae were fixed in 4% PFA/PBS pH 7.0 at room temperature for 1 h. Larvae were washed three times in 70% ethanol (EtOH), and 30% hydrochloric acid (HCl) for 5 min. Larvae were stained overnight in a solution of 0.1% alcian blue, 70% EtOH, and 30% HCl. Samples were then differentiated in 70% EtOH and 30% HCl until no blue color remained in the solution. For whole mount analysis, depigmented larvae were stepwise rehydrated to water and then cleared in a series of glycerol/1% KOH. For flat mount analysis, larvae were stepwise rehydrated to water and then rinsed three times in saturated sodium borate solution. Tissues were macerated in 3% trypsin in equal volumes of water and sodium borate for 15–30 min. Cartilages were dissected out using fine forceps and samples were rinsed in water before flat mounting.
Whole mount alcian stains were mounted laterally and ventrally in 3% methylcellulose in PBS and imaged on a Leica MZ FLIII fluorescent dissecting microscope connected to a computer running Leica FireCam image capture software (Leica Microsystems, Buffalo Grove, IL). Images were loaded into Adobe PhotoShop (Adobe Systems Inc., San Jose, CA) and cartilage length was measured for the bones that were not obstructed by tissue. Flat mounted cartilages were digitally photographed using a Zeiss Axioscope (Carl Zeiss Microscopy, LLC, One Zeiss Drive, Thornwood, NY 10594). The Cell Counter plugin of ImageJ105 was used to analyze and count cells in flat mount cartilages.
Acknowledgments
Lindsey McQuade, Ilya Shestopalov, and James K. Chen provided helpful advice. Part of the work was carried out using Core Technology Platform resources at New York University Abu Dhabi and mass spectrometry resources at the Proteomics and Mass Spectrometry Core Facility at the University of Georgia.
Glossary
Abbreviations
- GAD1
glutamic acid decarboxylase 1
- GAD2
glutamic acid decarboxylase 2
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.8b00231.
Preparation of BHQ- and CyHQ-based linkers 1a and 1b and CyHQ-linker-PEG (21), 1H NMR and 13C NMR spectra, UV–vis spectra, HPLC chromatograms, MS spectra; MO knockdown; enzymatic stability; Western blot analysis (PDF)
Author Contributions
M.J.O., L.L.B., D.D., R.E.B., A.T.P., A.J.V., K.T.H., S.P., and V.H. provided input into experimental design, conducted the experiments, interpreted the results, and assisted with writing the manuscript. M.J.O., L.L.B., and D.D. contributed equally. J.D.L. and T.M.D. conceived the overall idea, designed the experiments, interpreted the results, and wrote and edited the manuscript.
Supported by Grants R01 NS070159 from the National Institutes of Health to T.M.D. and J.D.L., R01 NS090645 from the National Institutes of Health to J.D.L., CHE-1012412 and CHE-1317760 from the National Science Foundation to T.M.D., and New York University Abu Dhabi.
The authors declare no competing financial interest.
Supplementary Material
References
- Brooks-Kayal A. R.; Raol Y. H.; Russek S. J. (2009) Alteration of epileptogenesis genes. Neurotherapeutics 6, 312–318. 10.1016/j.nurt.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortopan G. A.; Dinday M. T.; Baraban S. C. (2010) Spontaneous seizures and altered gene expression in GABA signaling pathways in a mind bomb mutant zebrafish. J. Neurosci. 30, 13718–13728. 10.1523/JNEUROSCI.1887-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash S. F.; Johnson R. S.; Tecott L. H.; Noebels J. L.; Mayfield R. D.; Hanahan D.; Baekkeskov S. (1997) Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. U. S. A. 94, 14060–14065. 10.1073/pnas.94.25.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S.; Huang H.-S. (2006) Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res. Rev. 52, 293–304. 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Guidotti A.; Auta J.; Davis J. M.; Gerevini V. D.; Dwivedi Y.; Grayson D. R.; Impagnatiello F.; Pandey G.; Pesold C.; Sharma R.; Uzunov D.; Costa E. (2000) Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder a postmortem brain study. Arch. Gen. Psychiatry 57, 1061–1069. 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Addington A. M.; Gornick M.; Duckworth J.; Sporn A.; Gogtay N.; Bobb A.; Greenstein D.; Lenane M.; Gochman P.; Baker N.; Balkissoon R.; Vakkalanka R. K.; Weinberger D. R.; Rapoport J. L.; Straub R. E. (2005) GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol. Psychiatry 10, 581–588. 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- Hossein Fatemi S.; Stary J. M.; Earle J. A.; Araghi-Niknam M.; Eagan E. (2005) GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr. Res. 72, 109–122. 10.1016/j.schres.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Fatemi S. H.; Halt A. R.; Stary J. M.; Kanodia R.; Schulz S. C.; Realmuto G. R. (2002) Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol. Psychiatry 52, 805–810. 10.1016/S0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- Lynex C. N.; Carr I. M.; Leek J. P.; Achuthan R.; Mitchell S.; Maher E. R.; Woods C. G.; Bonthon D. T.; Markham A. F. (2004) Homozygosity for a missense mutation in the 67 kDa isoform of glutamate decarboxylase in a family with autosomal recessive spastic cerebral palsy: parallels with Stiff-Person Syndrome and other movement disorders. BMC Neurol. 4, 20. 10.1186/1471-2377-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L. M.; Dalakas M. C.; Floeter M. K. (1999) The stiff-person syndrome: an autoimmune disorder affecting neurotransmission of gamma-aminobutyric acid. Ann. Intern. Med. 131, 522–530. 10.7326/0003-4819-131-7-199910050-00008. [DOI] [PubMed] [Google Scholar]
- Jessen K. R.; Hills J. M.; Saffrey M. J. (1986) Immunohistochemical demonstration of GABAergic neurons in the enteric nervous system. J. Neurosci. 6, 1628–1634. 10.1523/JNEUROSCI.06-06-01628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowery N. G.; Doble A.; Hill D. R.; Hudson A. L.; Shaw J. S.; Turnbull M. J.; Warrington R. (1981) Bicuculline-insensitive GABA receptors on peripheral autonomic nerve terminals. Eur. J. Pharmacol. 71, 53–70. 10.1016/0014-2999(81)90386-1. [DOI] [PubMed] [Google Scholar]
- Hill D. R.; Bowery N. G. (1981) 3H-baclofen and 3H-GABA bind to bicuculline-insensitive GABA B sites in rat brain. Nature 290, 149–152. 10.1038/290149a0. [DOI] [PubMed] [Google Scholar]
- Krantis A. (2000) GABA in the mammalian enteric nervous system. Physiology 15, 284–290. 10.1152/physiologyonline.2000.15.6.284. [DOI] [PubMed] [Google Scholar]
- Karlsen A. E.; Hagopian W. A.; Grubin C. E.; Dube S.; Disteche C. M.; Adler D. A.; Barmeier H.; Mathewes S.; Grant F. J.; et al (1991) Cloning and primary structure of a human islet isoform of glutamic acid decarboxylase from chromosome 10. Proc. Natl. Acad. Sci. U. S. A. 88, 8337–8341. 10.1073/pnas.88.19.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P.; Berggren P. O.; Bokvist K.; Ericson H.; Moehler H.; Oestenson C. G.; Smith P. A. (1989) Glucose-inhibition of glucagon secretion involves activation of GABAA-receptor chloride channels. Nature 341, 233–236. 10.1038/341233a0. [DOI] [PubMed] [Google Scholar]
- Xiang Y.-Y.; Wang S.; Liu M.; Hirota J. A.; Li J.; Ju W.; Fan Y.; Kelly M. M.; Ye B.; Orser B.; O’Byrne P. M.; Inman M. D.; Yang X.; Lu W.-Y. (2007) A GABAergic system in airway epithelium is essential for mucus overproduction in asthma. Nat. Med. 13, 862–867. 10.1038/nm1604. [DOI] [PubMed] [Google Scholar]
- Erdoe S. L.; Wolff J. R. (1990) γ-Aminobutyric acid outside the mammalian brain. J. Neurochem. 54, 363–372. 10.1111/j.1471-4159.1990.tb01882.x. [DOI] [PubMed] [Google Scholar]
- Baekkeskov S.; Aanstoot H. J.; Christgai S.; Reetz A.; Solimena M.; Cascalho M.; Folli F.; Richter-Olesen H.; Camilli P.-D. (1990) Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature 347, 151–156. 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- Kaufman D. L.; Erlander M. G.; Clare-Salzler M.; Atkinson M. A.; Maclaren N. K.; Tobin A. J. (1992) Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J. Clin. Invest. 89, 283–292. 10.1172/JCI115573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter W.; Endl J.; Eiermann T. H.; Brandt M.; Kientsch-Engel R.; Thivolet C.; Jungfer H.; Scherbaum W. A. (1992) Human monoclonal islet cell antibodies from a patient with insulin-dependent diabetes mellitus reveal glutamate decarboxylase as the target antigen. Proc. Natl. Acad. Sci. U. S. A. 89, 8467–8471. 10.1073/pnas.89.18.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander M. G.; Tillakaratne N. J. K.; Feldblum S.; Patel N.; Tobin A. J. (1991) Two genes encode distinct glutamate decarboxylases. Neuron 7, 91–100. 10.1016/0896-6273(91)90077-D. [DOI] [PubMed] [Google Scholar]
- Erlander M. G.; Tobin A. J. (1991) The structural and functional heterogeneity of glutamic acid decarboxylase: a review. Neurochem. Res. 16, 215–226. 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- Legay F.; Pelhate S.; Tappaz M. L. (1986) Phylogenesis of brain glutamic acid decarboxylase from vertebrates: immunochemical studies. J. Neurochem. 46, 1478–1486. 10.1111/j.1471-4159.1986.tb01765.x. [DOI] [PubMed] [Google Scholar]
- Martin S. C.; Heinrich G.; Sandell J. H. (1998) Sequence and expression of glutamic acid decarboxylase isoforms in the developing zebrafish. J. Comp. Neurol. 396, 253–266. . [DOI] [PubMed] [Google Scholar]
- Martin D. L.; Liu H.; Martin S. B.; Wu S. J. (2000) Structural features and regulatory properties of the brain glutamate decarboxylases. Neurochem. Int. 37, 111–119. 10.1016/S0197-0186(00)00014-0. [DOI] [PubMed] [Google Scholar]
- Kaufman D. L.; Houser C. R.; Tobin A. J. (1991) Two forms of the γ-aminobutyric acid synthetic enzyme glutamate decarboxylase have distinct intraneuronal distributions and cofactor interactions. J. Neurochem. 56, 720–723. 10.1111/j.1471-4159.1991.tb08211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal C. S.; Tobin A. J. (1998) Uniqueness and redundancy in GABA production. Perspect. Dev. Neurobiol. 5, 109–118. [PubMed] [Google Scholar]
- Esclapez M.; Tillakaratne N. J. K.; Tobin A. J.; Houser C. R. (1993) Comparative localization of mRNAs encoding two forms of glutamic acid decarboxylase with nonradioactive in situ hybridization methods. J. Comp. Neurol. 331, 339–362. 10.1002/cne.903310305. [DOI] [PubMed] [Google Scholar]
- Feldblum S.; Erlander M. G.; Tobin A. J. (1993) Different distributions of GAD65 and GAD67 mRNAs suggest that the two glutamate decarboxylases play distinctive functional roles. J. Neurosci. Res. 34, 689–706. 10.1002/jnr.490340612. [DOI] [PubMed] [Google Scholar]
- Sheikh S. N.; Martin S. B.; Martin D. L. (1999) Regional distribution and relative amounts of glutamate decarboxylase isoforms in rat and mouse brain. Neurochem. Int. 35, 73–80. 10.1016/S0197-0186(99)00063-7. [DOI] [PubMed] [Google Scholar]
- Cocco A.; Roennberg A. M. C.; Jin Z.; Andre G. I.; Vossen L. E.; Bhandage A. K.; Thoernqvist P.-O.; Birnir B.; Winberg S. (2017) Characterization of the γ-aminobutyric acid signaling system in the zebrafish (Danio rerio Hamilton) central nervous system by reverse transcription-quantitative polymerase chain reaction. Neuroscience 343, 300–321. 10.1016/j.neuroscience.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Varju P.; Katarova Z.; Madarasz E.; Szabo G. (2001) GABA signalling during development: new data and old questions. Cell Tissue Res. 305, 239–246. 10.1007/s004410100356. [DOI] [PubMed] [Google Scholar]
- Katarova Z.; Sekerkova G.; Prodan S.; Mugnaini E.; Szabo G. (2000) Domain-restricted expression of two glutamic acid decarboxylase genes in midgestation mouse embryos. J. Comp. Neurol. 424, 607–627. . [DOI] [PubMed] [Google Scholar]
- Condie B. G.; Bain G.; Gottlieb D. I.; Capecchi M. R. (1997) Cleft palate in mice with a targeted mutation in the γ-aminobutyric acid-producing enzyme glutamic acid decarboxylase 67. Proc. Natl. Acad. Sci. U. S. A. 94, 11451–11455. 10.1073/pnas.94.21.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada H.; Kawamura Y.; Maruyama K.; Kume H.; Ding R.-G.; Kanbara N.; Kuzume H.; Sanbo M.; Yagi T.; Obata K. (1997) Cleft palate and decreased brain γ-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. U. S. A. 94, 6496–6499. 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culiat C. T.; Stubbs L. J.; Woychik R. P.; Russell L. B.; Johnson D. K.; Rinchik E. M. (1995) Deficiency of the β3 subunit of the type A γ-aminobutyric acid receptor causes cleft palate in mice. Nat. Genet. 11, 344–346. 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- Homanics G. E.; DeLorey T. M.; Firestone L. L.; Quinlan J. J.; Handforth A.; Harrison N. L.; Krasowski M. D.; Rick C. E.; Korpi E. R.; Makela R.; Brilliant M. H.; Hagiwara N.; Ferguson C.; Snyder K.; Olsen R. W. (1997) Mice devoid of gamma-aminobutyrate type A receptor beta3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proc. Natl. Acad. Sci. U. S. A. 94, 4143–4148. 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno K.; Suzuki Y.; Yamada A.; Aoki Y.; Kure S.; Matsubara Y. (2004) Association between nonsyndromic cleft lip with or without cleft palate and the glutamic acid decarboxylase 67 gene in the Japanese population. Am. J. Med. Genet. 127A, 11–16. 10.1002/ajmg.a.20649. [DOI] [PubMed] [Google Scholar]
- Scapoli L.; Martinelli M.; Pezzetti F.; Carinci F.; Bodo M.; Tognon M.; Carinci P. (2002) Linkage disequilibrium between GABRB3 gene and nonsyndromic familial cleft lip with or without cleft palate. Hum. Genet. 110, 15–20. 10.1007/s00439-001-0639-5. [DOI] [PubMed] [Google Scholar]
- Deiters A.; Yoder J. A. (2006) Conditional Transgene and Gene Targeting Methodologies in Zebrafish. Zebrafish 3, 415–429. 10.1089/zeb.2006.3.415. [DOI] [PubMed] [Google Scholar]
- Esengil H.; Chen J. K. (2008) Gene regulation technologies in zebrafish. Mol. BioSyst. 4, 300–308. 10.1039/b718447f. [DOI] [PubMed] [Google Scholar]
- Shestopalov I. A.; Chen J. K. (2008) Chemical technologies for probing embryonic development. Chem. Soc. Rev. 37, 1294–1307. 10.1039/b703023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A.; Ekker S. C. (2000) Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26, 216–220. 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Skromne I.; Prince V. E. (2008) Current perspectives in zebrafish reverse genetics: moving forward. Dev. Dyn. 237, 861–882. 10.1002/dvdy.21484. [DOI] [PubMed] [Google Scholar]
- Tang X.; Maegawa S.; Weinberg E. S.; Dmochowski I. J. (2007) Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 129, 11000–11001. 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- Dmochowski I. J.; Tang X. (2007) Taking control of gene expression with light-activated oligonucleotides. BioTechniques 43, 161. 10.2144/000112519. [DOI] [PubMed] [Google Scholar]; 163,165,167,169,171
- Tang X.; Dmochowski I. J. (2007) Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 3, 100–110. 10.1039/B614349K. [DOI] [PubMed] [Google Scholar]
- Tang X.; Dmochowski I. J. (2006) Controlling RNA digestion by RNase H with a light-activated DNA hairpin. Angew. Chem., Int. Ed. 45, 3523–3526. 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]
- Shestopalov I. A.; Sinha S.; Chen J. K. (2007) Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 3, 650–651. 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- Ouyang X.; Shestopalov I. A.; Sinha S.; Zheng G.; Pitt C. L. W.; Li W.-H.; Olson A. J.; Chen J. K. (2009) Versatile Synthesis and Rational Design of Caged Morpholinos. J. Am. Chem. Soc. 131, 13255–13269. 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoryak O. D.; Dore T. M. (2002) Brominated hydroxyquinoline as a photolabile protecting group with sensitivity to multiphoton excitation. Org. Lett. 4, 3419–3422. 10.1021/ol026524g. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Pavlos C. M.; Toscano J. P.; Dore T. M. (2006) 8-Bromo-7-hydroxyquinoline as a Photoremovable Protecting Group for Physiological Use: Mechanism and Scope. J. Am. Chem. Soc. 128, 4267–4276. 10.1021/ja0555320. [DOI] [PubMed] [Google Scholar]
- Tomasini A. J.; Schuler A. D.; Zebala J. A.; Mayer A. N. (2009) PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis 47, 736–743. 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallafuss A.; Gibson D.; Morcos P.; Li Y.; Seredick S.; Eisen J.; Washbourne P. (2012) Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development 139, 1691–1699. 10.1242/dev.072702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A.; Garner R. A.; Lusic H.; Govan J. M.; Dush M.; Nascone-Yoder N. M.; Yoder J. A. (2010) Photocaged Morpholino Oligomers for the Light-Regulation of Gene Function in Zebrafish and Xenopus Embryos. J. Am. Chem. Soc. 132, 15644–15650. 10.1021/ja1053863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe S.; Shestopalov I. A.; Provost E.; Leach S. D.; Chen J. K. (2012) Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem., Int. Ed. 51, 6908–6911. 10.1002/anie.201201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wu L.; Wang P.; Lv C.; Yang Z.; Tang X. (2012) Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res. 40, 11155–11162. 10.1093/nar/gks840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe S.; Liu Q.; McQuade L. E.; Deiters A.; Chen J. K. (2014) Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem., Int. Ed. 53, 10114–10118. 10.1002/anie.201405355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griepenburg J. C.; Rapp T. L.; Carroll P. J.; Eberwine J.; Dmochowski I. J. (2015) Ruthenium-caged antisense morpholinos for regulating gene expression in zebrafish embryos. Chem. Sci. 6, 2342–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. J.; Kragor C. H.; Reddie K. G.; Wilson H. C.; Zhu Y.; Dore T. M. (2009) Substituent Effects on the Sensitivity of a Quinoline Photoremovable Protecting Group to One- and Two-Photon Excitation. J. Org. Chem. 74, 1721–1729. 10.1021/jo802658a. [DOI] [PubMed] [Google Scholar]
- Grone B. P.; Maruska K. P. (2016) Three Distinct Glutamate Decarboxylase Genes in Vertebrates. Sci. Rep. 6, 30507. 10.1038/srep30507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.; Fagernes C. E.; Nilsson G. E.; Jutfelt F. (2016) Expression of genes involved in brain GABAergic neurotransmission in three-spined stickleback exposed to near-future CO2. Conserv. Physiol. 4, cow068. 10.1093/conphys/cow068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.; Fagernes C. E.; Jutfelt F.; Nilsson G. E. (2017) Erratum: Expression of genes involved in brain GABAergic neurotransmission in three-spined stickleback exposed to near-future CO2. Conserv. Physiol. 5, cox004. 10.1093/conphys/cox004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald R. B.; Debiais-Thibaud M.; Talbot J. C.; Ekker M. (2010) The relationship between dlx and gad1 expression indicates highly conserved genetic pathways in the zebrafish forebrain. Dev. Dyn. 239, 2298–2306. 10.1002/dvdy.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S.-i.; Mandel G.; Fetcho J. R. (2004) Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J. Comp. Neurol. 480, 1–18. 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- Higashijima S.-i.; Schaefer M.; Fetcho J. R. (2004) Neurotransmitter properties of spinal interneurons in embryonic and larval zebrafish. J. Comp. Neurol. 480, 19–37. 10.1002/cne.20279. [DOI] [PubMed] [Google Scholar]
- Asad N.; Deodato D.; Lan X.; Widegren M. B.; Phillips D. L.; Du L.; Dore T. M. (2017) Photochemical Activation of Tertiary Amines for Applications in Studying Cell Physiology. J. Am. Chem. Soc. 139, 12591–12600. 10.1021/jacs.7b06363. [DOI] [PubMed] [Google Scholar]
- Asada H.; Kawamura Y.; Maruyama K.; Kume K.; Ding R.-g.; Ji F. Y.; Kanbara N.; Kuzume H.; Sanbo M.; Yagi T.; Obata K. (1996) Mice lacking the 65 kDa isoform of glutamic acid decarboxylase (GAD65) maintain normal levels of GAD67 and GABA in their brains but are susceptible to seizures. Biochem. Biophys. Res. Commun. 229, 891–895. 10.1006/bbrc.1996.1898. [DOI] [PubMed] [Google Scholar]
- Eisen J. S.; Smith J. C. (2008) Controlling morpholino experiments: don’t stop making antisense. Development 135, 1735–1743. 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Stainier D. Y. R.; Raz E.; Lawson N. D.; Ekker S. C.; Burdine R. D.; Eisen J. S.; Ingham P. W.; Schulte-Merker S.; Yelon D.; Weinstein B. M.; Mullins M. C.; Wilson S. W.; Ramakrishnan L.; Amacher S. L.; Neuhauss S. C. F.; Meng A.; Mochizuki N.; Panula P.; Moens C. B. (2017) Guidelines for morpholino use in zebrafish. PLoS Genet. 13, e1007000. 10.1371/journal.pgen.1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris M.; Schloesser M.; Baurain D.; Hanikenne M.; Muller M.; Motte P. (2017) Number of inadvertent RNA targets for morpholino knockdown in Danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors. Nucleic Acids Res. 45, 9547–9557. 10.1093/nar/gkx638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R.; Tsunekawa N.; Obata K. (2004) Cleft palate by picrotoxin or 3-MP and palatal shelf elevation in GABA-deficient mice. Neurotoxicol. Teratol. 26, 587–592. 10.1016/j.ntt.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Hagiwara N.; Katarova Z.; Siracusa L. D.; Brilliant M. H. (2003) Nonneuronal expression of the GABA(A) beta3 subunit gene is required for normal palate development in mice. Dev. Biol. 254, 93–101. 10.1016/S0012-1606(02)00030-1. [DOI] [PubMed] [Google Scholar]
- Zimmerman E. F.; Wee E. L. (1984) Role of neurotransmitters in palate development. Curr. Top. Dev. Biol. 19, 37–63. 10.1016/S0070-2153(08)60394-4. [DOI] [PubMed] [Google Scholar]
- Wee E. L.; Norman E. J.; Zimmerman E. F. (1986) Presence of γ-aminobutyric acid in embryonic palates of AJ and SWV mouse strains. J. Craniofacial Genet. Dev. Biol. 6, 53–61. [PubMed] [Google Scholar]
- Culiat C. T.; Stubbs L. J.; Woychik R. P.; Russell L. B.; Johnson D. K.; Rinchik E. M. (1995) Deficiency of the beta 3 subunit of the type A gamma-aminobutyric acid receptor causes cleft palate in mice. Nat. Genet. 11, 344–346. 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- Miller R. P.; Becker B. A. (1975) Teratogenicity of oral diazepam and diphenylhydantoin in mice. Toxicol. Appl. Pharmacol. 32, 53–61. 10.1016/0041-008X(75)90194-5. [DOI] [PubMed] [Google Scholar]
- Jurand A.; Martin L. V. H. (1994) Cleft palate and open eyelids inducing activity of lorazepam and the effect of flumazenil, the benzodiazepine antagonist. Pharmacol. Toxicol. 74, 228–235. 10.1111/j.1600-0773.1994.tb01103.x. [DOI] [PubMed] [Google Scholar]
- Wee E. L.; Zimmerman E. F. (1983) Involvement of GABA in palate morphogenesis and its relation to diazepam teratogenesis in two mouse strains. Teratology 28, 15–22. 10.1002/tera.1420280104. [DOI] [PubMed] [Google Scholar]
- Aarskog D. (1975) Association between maternal intake of diazepam and oral clefts. Lancet 306, 921. 10.1016/S0140-6736(75)92153-4. [DOI] [PubMed] [Google Scholar]
- Safra M. J.; Oakley G. P. Jr. (1975) Association between cleft lip with or without cleft palate and prenatal exposure to diazepam. Lancet 306, 478–480. 10.1016/S0140-6736(75)90548-6. [DOI] [PubMed] [Google Scholar]
- Saxen I.; Saxen L. (1975) Association between maternal intake of diazepam and oral clefts. Lancet 306, 498. 10.1016/S0140-6736(75)90567-X. [DOI] [PubMed] [Google Scholar]
- Rosenberg L.; Mitchell A. A.; Parsells J. L.; Pashayan H.; Louik C.; Shapiro S. (1983) Lack of relation of oral clefts to diazepam use during pregnancy. N. Engl. J. Med. 309, 1282–1285. 10.1056/NEJM198311243092103. [DOI] [PubMed] [Google Scholar]
- Loescher W. (1979) 3-Mercaptopropionic acid: convulsant properties, effects on enzymes of the γ-aminobutyrate system in mouse brain and antagonism by certain anticonvulsant drugs, aminooxyacetic acid and gabaculine. Biochem. Pharmacol. 28, 1397–1407. 10.1016/0006-2952(79)90443-X. [DOI] [PubMed] [Google Scholar]
- Rodriguez de Lores Arnaiz G.; Alberici de Canal M.; Robiolo B.; Mistrorigo de Pacheco M. (1973) Effect of the convulsant 3-mercaptopropionic acid on enzymes of the γ-aminobutyrate system in the rat cerebral cortex. J. Neurochem. 21, 615–623. 10.1111/j.1471-4159.1973.tb06006.x. [DOI] [PubMed] [Google Scholar]
- Leweke F. M.; Louvel J.; Rausche G.; Heinemann U. (1990) Effects of pentetrazol on neuronal activity and on extracellular calcium concentration in rat hippocampal slices. Epilepsy Res. 6, 187–198. 10.1016/0920-1211(90)90072-4. [DOI] [PubMed] [Google Scholar]
- Baraban S. C.; Taylor M. R.; Castro P. A.; Baier H. (2005) Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience 131, 759–768. 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Tao L.; Lauderdale J. D.; Sornborger A. T. (2011) Mapping Functional Connectivity between Neuronal Ensembles with Larval Zebrafish Transgenic for a Ratiometric Calcium Indicator. Front. Neural Circuits 5, 2. 10.3389/fncir.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A. (2012) Pentylenetetrazol (PTZ) kindling model of epilepsy. Curr. Protoc. Neurosci. 58, 9.37.31–39.37.12. 10.1002/0471142301.ns0937s58. [DOI] [PubMed] [Google Scholar]
- Hewapathirane D. S.; Dunfield D.; Yen W.; Chen S.; Haas K. (2008) In vivo imaging of seizure activity in a novel developmental seizure model. Exp. Neurol. 211, 480–488. 10.1016/j.expneurol.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Livingston R. (1971) Behavior of Photochromic Systems. In Photochromism (Brown G. H., Ed.), pp 13–44, Wiley, New York. [Google Scholar]
- Hatchard C. G.; Parker C. A. (1956) A new sensitive chemical actinometer. II. Potassium ferrioxalate as a standard chemical actinometer. Proc. R. Soc. London, Ser. A 235, 518–536. 10.1098/rspa.1956.0102. [DOI] [Google Scholar]
- Westerfield M., Ed. (2007) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th ed., University of Oregon Press, Eugene, OR. [Google Scholar]
- Kimmel C. B.; Ballard W. W.; Kimmel S. R.; Ullmann B.; Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Wloga D.; Dougan S. T. (2011) Embryological manipulations in zebrafish. Methods Mol. Biol. 770, 139–184. 10.1007/978-1-61779-210-6_6. [DOI] [PubMed] [Google Scholar]
- Bill B. R.; Petzold A. M.; Clark K. J.; Schimmenti L. A.; Ekker S. C. (2009) A Primer for Morpholino Use in Zebrafish. Zebrafish 6, 69–77. 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutkiewicz E. M.; Baladi M. G.; Folk J. E.; Rice K. C.; Woods J. H. (2006) The convulsive and electroencephalographic changes produced by nonpeptidic δ-opioid agonists in rats: comparison with pentylenetetrazol. J. Pharmacol. Exp. Ther. 317, 1337–1348. 10.1124/jpet.105.095810. [DOI] [PubMed] [Google Scholar]
- Peterson R. T.; Fishman M. C. (2004) Discovery and use of small molecules for probing biological processes in zebrafish. Methods Cell Biol. 76, 569–591. 10.1016/S0091-679X(04)76026-4. [DOI] [PubMed] [Google Scholar]
- Trapani J. G.; Nicolson T. (2010) Physiological recordings from zebrafish lateral-line hair cells and afferent neurons. Methods Cell Biol. 100, 219–231. 10.1016/B978-0-12-384892-5.00008-6. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Dahm R., Eds. (2002) Zebrafish: A Practical Approach, Oxford University Press, Oxford, UK. [Google Scholar]
- Harlow E., and Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B.; Miller C. T.; Kruze G.; Ullmann B.; BreMiller R. A.; Larison K. D.; Snyder H. C. (1998) The Shaping of Pharyngeal Cartilages during Early Development of the Zebrafish. Dev. Biol. 203, 245–263. 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.