Fig. 9.
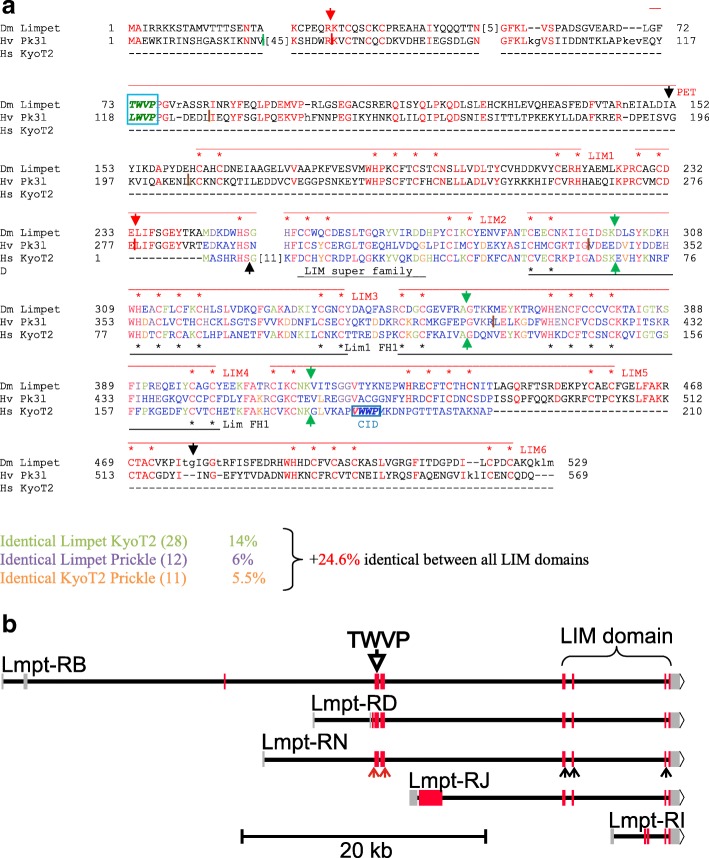
Alignment of fly Limpet, human KyoT2 and Hydra Prickle 3 like. a The alignment shows good conservation in the LIM domain between all three proteins. Best identity score in the LIM domain is between KyoT2 and Limpet (38.6%), followed by Limpet and Prickle 3 like (30.6%) and KyoT2 and Prickle 3 like (30.1%). Limpet and Prickle 3 like share considerable conservation over the entire length; the TWVP tetra-peptide motif is present at the identical position in the PET domain (italic green, boxed). Moreover, the two share conserved intron positions (red arrows and dashes). Green arrows indicate identical intron positions between Limpet and KyoT2. Black arrows and dashes mark intron positions specific to Hydra Prickle 3 like. Red residues are identical in all three proteins, green identical between Limpet and KyoT2, pink identical between Limpet and Prickle 3 like, and orange identical between KyoT2 and Prickle 3 like, blue are not conserved. b Scheme of the genomic organization of the D. melanogaster Limpet gene with representative transcripts (adapted from flybase). 15 transcripts are predicted, however, with 8 different open reading frames that fall into three classes: One encoding PET and LIM domain proteins (RB, RD and RN are shown as examples), one encoding LIM domain only proteins (RI, and RJ are shown as example), and the third containing neither (not shown). The exon encoding the PET domain with the TWVP tetra-peptide is indicated by the open arrow. The small downstream exons encode the LIM domain. Red arrows indicate exon/intron boundaries conserved between Limpet and Prickle 3 like, black arrows indicate exon/intron boundaries conserved between Limpet and KyoT2