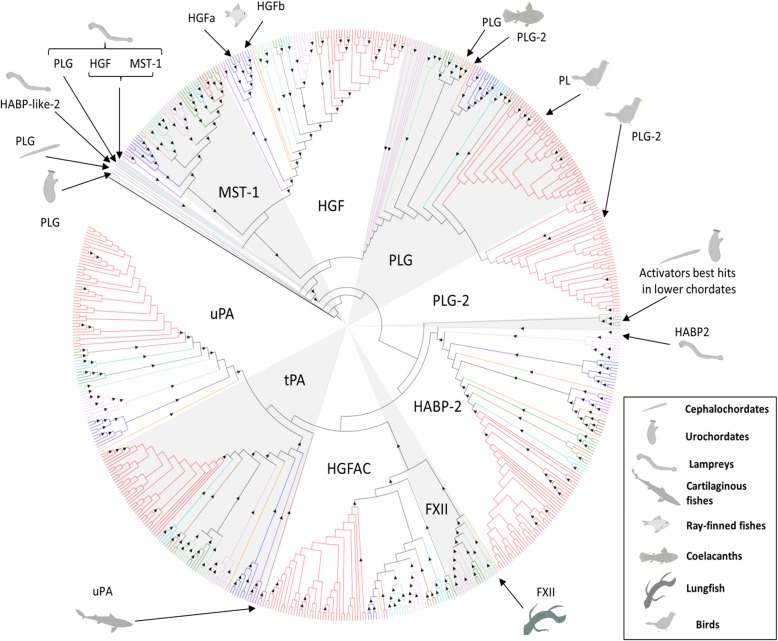
Fig. 4.
Maximum-likelihood phylogenetic tree of the PLG and PLG activator groups of serine proteases. Caenorhabditis elegans SVH-1 (BAL45941.1) was chosen as outgroup to root the tree. Triangles within branches represent bootstrap support higher than 50% after 1000 replicates. Colored branches indicate lower chordates (grey), cartilaginous fishes (violet), ray-finned fishes (blue), coelacanths and lungfish (orange), amphibians (light green), lizards and snakes (dark green), turtles (turquoise), crocodilians (brown) and birds (red). Position of the non-canonical or first appeared candidates is indicated within the tree by arrows. PLG-2 corresponds to the extra PLG gene identified, which has lost the catalytic activity. PLG (plasminogen), HGF (hepatocyte growth factor), MST-1 (macrophage stimulating 1), HABP2 (hyaluronan binding protein 2), HGFAC (hepatocyte growth factor activator), tPA (tissue-type plasminogen activator), uPA (urokinase-type plasminogen activator) and FXII (coagulation factor XII/Hageman factor). The unrooted tree with bootstrap values is provided in Additional file 3