Abstract
Giardia lamblia is a binucleate protistan parasite causing significant diarrheal disease worldwide. An inability to target Cas9 to both nuclei, combined with the lack of nonhomologous end joining and markers for positive selection, has stalled the adaptation of CRISPR/Cas9-mediated genetic tools for this widespread parasite. CRISPR interference (CRISPRi) is a modification of the CRISPR/Cas9 system that directs catalytically inactive Cas9 (dCas9) to target loci for stable transcriptional repression. Using a Giardia nuclear localization signal to target dCas9 to both nuclei, we developed efficient and stable CRISPRi-mediated transcriptional repression of exogenous and endogenous genes in Giardia. Specifically, CRISPRi knockdown of kinesin-2a and kinesin-13 causes severe flagellar length defects that mirror defects with morpholino knockdown. Knockdown of the ventral disk MBP protein also causes severe structural defects that are highly prevalent and persist in the population more than 5 d longer than defects associated with transient morpholino-based knockdown. By expressing two guide RNAs in tandem to simultaneously knock down kinesin-13 and MBP, we created a stable dual knockdown strain with both flagellar length and disk defects. The efficiency and simplicity of CRISPRi in polyploid Giardia allows rapid evaluation of knockdown phenotypes and highlights the utility of CRISPRi for emerging model systems.
INTRODUCTION
Giardia lamblia is a common parasitic protist that infects more than 300 million people worldwide each year, causing acute diarrheal disease in areas with inadequate sanitation and water treatment (Einarsson et al., 2016). Giardia has a two-stage life cycle in which mammalian hosts ingest quadrinucleate cysts from contaminated water sources. As they pass into the gastrointestinal tract, cysts develop into binucleate, flagellated trophozoites. During infection, trophozoites attach extracellularly to the gut epithelium, proliferate, and later differentiate back into infectious cysts that are passed into the environment (Heyworth, 2014). Giardiasis is considered a neglected disease (Savioli et al., 2006), and the need for new effective and affordable treatments for giardiasis is underscored by estimated failure rates of up to 20% for standard treatments (Upcroft and Upcroft, 2001), emerging evidence of drug resistance (Upcroft et al., 1996; Barat and Bloland, 1997; Land and Johnson, 1999), and the prevalence of chronic or recurrent infections (Bartelt and Sartor, 2015).
The nuclei in trophozoites and cysts are diploid (Bernander et al., 2001), transcriptionally active (Kabnick and Peattie, 1990), and genetically equivalent (Morrison et al., 2007; Franzen et al., 2009; Adam et al., 2013; Hanevik et al., 2015). Because Giardia trophozoites are effectively tetraploid and lack a defined sexual cycle (Poxleitner et al., 2008), the development of molecular genetic tools in Giardia has generally lagged behind that of other parasitic protists. Although new CRISPR/Cas9 gene-editing strategies have revolutionized molecular genetics in other protists (Grzybek et al., 2018), the adaptation of CRISPR-based tools for Giardia has been hindered by Giardia's lack of a nonhomologous end-joining (NHEJ) pathway (Morrison et al., 2007) and by the inability to target native Streptomyces pyogenes Cas9 (SpCas9) to the two nuclei. The widely used SV40 nuclear localizing signal (NLS) failed to localize full-length Cas9 (Ebneter et al., 2016), although it has been used previously in Giardia to localize exogenously expressed GFP or TetR protein to the nuclei (Elmendorf et al., 2000). For these reasons, the first Giardia quadruple knockout strain of the cyst wall component CWP1 was constructed using Cre/loxP sequential gene disruption, rather than a CRISPR/Cas9-mediated strategy (Ebneter et al., 2016).
Gene knockouts are only one of many strategies for interrogating gene function (Qi et al., 2013), and null alleles in essential processes such as cell cycle regulation (Horlock-Roberts et al., 2017) and cytokinesis (Hardin et al., 2017) can result in severely reduced fitness or in lethality. Translational repression using morpholino oligonucleotides has been used extensively in Giardia (House et al., 2011; Paredez et al., 2011; Woessner and Dawson, 2012), but morpholinos are transient (lasting less than 48 h), lack complete penetrance, and are costly, making morpholino knockdowns less useful for genome-wide functional screens (Krtkova and Paredez, 2017). Both the transience and incomplete penetrance of morpholino knockdowns are problematic for evaluation of infection dynamics of Giardia mutants (Barash et al., 2017). Transcriptional repression is an important alternative strategy for the characterization of gene function; however, despite the presence of conserved components of the RNA interference (RNAi) machinery, RNAi does not efficiently silence genes in Giardia (Krtkova and Paredez, 2017).
CRISPR/Cas9 is a modular and flexible DNA-binding platform that has been adapted for applications beyond genome editing, including transcriptional repression (Larson et al., 2013). The CRISPR interference system (CRISPRi) is a modification of the CRISPR/Cas9 system, in which a catalytically inactive or “dead” Cas9 (dCas9) induces stable, inducible, or reversible gene knockdown in eukaryotes (Larson et al., 2013; Piatek et al., 2015), as well as diverse bacteria (Larson et al., 2013; Zhang et al., 2016; Liu et al., 2017; Tao et al., 2017; Zuberi et al., 2017; Kaczmarzyk et al., 2018). Using a complementary guide RNA (gRNA), the inactive dCas9 is directed to precise genomic targets, where it binds and inhibits transcription initiation and/or elongation rather than inducing double-stranded breaks in DNA (Larson et al., 2013). CRISPRi has significant advantages over RNAi or morpholinos, as it directly and stably inhibits transcription (Larson et al., 2013). In many systems, CRISPRi is empirically as effective as RNAi in transcriptional silencing, with significantly fewer off-target effects (Larson et al., 2013).
Here we demonstrate precise and stable CRISPRi-mediated transcriptional repression of both exogenous and endogenous genes in Giardia. This first successful application of CRISPRi for transcriptional knockdown in a parasitic protist—or a polyploid eukaryote—highlights the utility of CRISPRi for emerging model systems. Using a Giardia nuclear localization signal (NLS) to target dCas9 to both nuclei, we show efficient and persistent CRISPRi-mediated repression of one exogenous reporter gene and three endogenous Giardia cytoskeletal genes. CRISPRi knockdowns have severe cytoskeletal phenotypes that are highly prevalent and persist at least 1 wk in cultured trophozoites. We also tandemly express gRNAs to simultaneously knock down one flagellar and one ventral disk gene, resulting in a stable strain with two knockdown phenotypes. The efficiency and simplicity of CRISPRi in Giardia permits the rapid assessment of the phenotypic consequences of protein knockdown (Larson et al., 2013; Jost et al., 2017) and will likely enable the first forward and reverse genetic screens.
RESULTS
The SV40 NLS is not sufficient to localize Cas9 to the two Giardia nuclei
To express Cas9 in Giardia, we cloned mammalian codon–optimized S. pyogenes Cas9 (mCas9) with a C-terminal SV40 nuclear localization signal (NLS) and a 3XHA epitope tag into the pcGFP1Fpac backbone (Hagen et al., 2011), creating a Cas9-SV40NLS-3XHA-GFP fusion (Figure 1A). A comparison of the modal codon usage frequencies (Davis and Olsen, 2010) of mCas9 and all protein-coding genes of G. lamblia ATCC 50803 indicated that further codon optimization of mCas9 was unnecessary. As Cas9 toxicity has been reported in some systems (Jiang et al., 2014), Cas9 was expressed using a moderate-strength Giardia promoter for the constitutive malate dehydrogenase (MDH, GL50803_3331) gene (Figure 1A).
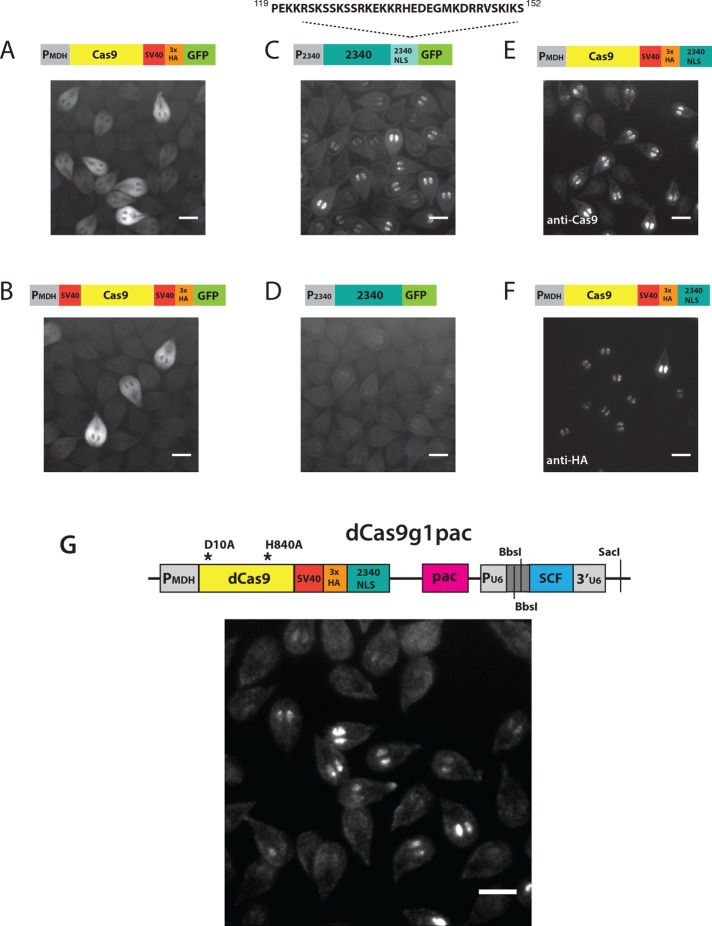
FIGURE 1:
A Giardia-specific NLS is necessary and sufficient to localize Cas9 and dCas9 to both nuclei. C-terminal GFP-tagged Cas9 with either a single C-terminal SV40 NLS (A) or dual N- and C-terminal SV40 NLSs (B) localizes only to the cytoplasm. The presence of a Giardia-specific native 34–amino acid C-terminal NLS from the Giardia protein GL50803_2340 (2340NLS) is necessary for the localization of 2340GFP to both nuclei (C, D). The C-terminal addition of the 2340NLS to Cas9 is sufficient for localization to both nuclei, as shown by immunostaining with anti-Cas9 (E) or anti-HA (F). A schematic of the Giardia CRISPRi vector dCas9g1pac and the localization of dCas9 as determined by anti-Cas9 antibody staining are shown in G. The vector includes catalytically inactive dCas9 with a C-terminal 2340NLS and a 3XHA epitope tag driven by the Giardia malate dehydrogenase promoter (PMDH) and a puromycin resistance marker (pac) for positive selection in Giardia. The gRNA cassette is expressed using the Giardia U6 spliceosomal RNA pol III promoter and includes inverted BbsI restriction sites for rapid cloning of specific gRNA target sequences, followed by the gRNA scaffold sequence (SCF). Additional gRNA cassettes are added at the SacI site (Materials and Methods). Anti-Cas9 immunostaining indicates over 50% of cells express dCas9 in both nuclei. All scale bars = 5 µm.
The Cas9-GFP fusion with a C-terminal SV40 NLS localized only to the cytoplasm, with no signal detected in either of the nuclei (Figure 1A). Cas9 localization to the nucleus is required for Cas9 genome editing; therefore we added a second SV40 NLS at the N-terminus. However, addition of another SV40 NLS had no impact on nuclear localization (Figure 1B). Thus, the SV40 NLS, which is commonly used in other systems and for recombinant Cas9 in commercial Cas9/CRISPR kits, is not sufficient for nuclear localization of Cas9 in Giardia. The failure of the SV40 NLS to target full-length Cas9 to Giardia's nuclei has been noted recently (Ebneter et al., 2016), although a truncated and inactive Cas9 was successfully localized with this NLS (Ebneter et al., 2016), as were GFP and TetR (Elmendorf et al., 2000).
Defining a native Giardia NLS to target Cas9 and dCas9 to both nuclei
To localize Cas9 to Giardia's nuclei, we used the NLS prediction software NLStradamus to screen for putative NLS sequences (Nguyen Ba et al., 2009). We identified 13 putative NLSs in 65 proteins with known nuclear localization (Figure 1C and Supplemental Material). To determine whether the putative NLSs were necessary for nuclear localization, we deleted C-terminal putative NLS sequences from seven nuclear-localizing GFP-tagged Giardia proteins and assayed for loss of nuclear localization. Deletion of putative NLSs from three proteins resulted in a loss of nuclear localization or an increase in localization to the cytoplasm (Figure 1D and Supplemental Material). Deletion of the other four NLSs had no effect on nuclear localization. All three candidate NLSs were sufficient for Cas9 localization to the nuclei (Supplemental Material). Cas9 nuclear localization was most intense and prevalent with the 34-amino acid C-terminal NLS from the Giardia protein GL50803_2340 (Figure 1, E and F). Using the 2340NLS fused to the C-terminus of dCas9, we created a Giardia CRISPR interference expression vector (dCas9g1pac) that included a gRNA cassette with a Giardia U6 promoter and puromycin (pac) selection (Materials and Methods and Figure 1G).
CRISPRi-mediated knockdown of the exogenous NanoLuc reporter gene
To assess the ability of CRISPRi to repress transcription in Giardia, we created a strain that constitutively expresses the NanoLuc (NLuc) reporter (Hall et al., 2012) from the Giardia MDH promoter on a plasmid with neomycin selection. We designed eight gRNAs (+53, +147, +193, +255, +285, +365, +444, +488) (Figure 2A) to target the entire coding region of the NLuc gene at ∼50–base pair intervals. One gRNA (-11) was designed to target just upstream of the translation start site. Seven of nine gRNAs significantly repressed NLuc luminescence by ∼20–61% as compared with the NanoLucNeo strain (Figure 2B).
FIGURE 2:
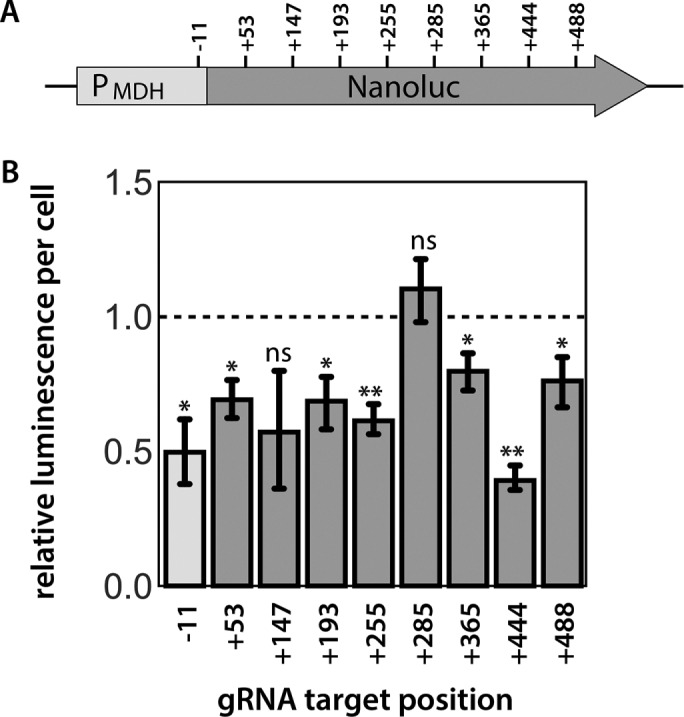
CRISPRi-mediated knockdown of exogenously expressed NanoLuc in Giardia. Nine gRNAs were designed to repress transcription of the luciferase reporter gene NanoLuc expressed exogenously from the Giardia MDH promoter (A). Eight gRNAs targeted the coding region of the NanoLuc gene and one gRNA (-11) targeted the region immediately upstream of the translation start site. Target positions are numbered relative to the translational start site ATG and are located three bases upstream of the PAM sequence (NGG) for each gRNA. Relative luminescence per cell is plotted for each gRNA designed to repress Nanoluc expression. In B, the mean luminescence per cell for two independent transformations is compared between the different luciferase knockdown strains. Error bars indicate 95% confidence intervals. The significance of luciferase knockdown relative to a nonspecific gRNA control was assessed using an unpaired t test, with ns = not significant, *p ≤ 0.05, **p ≤ 0.01 (Materials and Methods).
CRISPRi-mediated knockdowns of kinesin-13 or kinesin-2a cause significant alterations in flagellar length
Giardia has four pairs of bilaterally symmetric flagella that are maintained at consistent equilibrium lengths (Figure 3A). Giardia possesses a single heterotrimeric kinesin-2 motor, consisting of kinesin-2a and -2b and the kinesin-associated protein (KAP) that is required for flagellar assembly and maintenance (Hoeng et al., 2008). The length of a flagellum is determined by the balance of kinesin-2 dependent assembly (Kozminski et al., 1995) and kinesin-13 dependent disassembly at the distal flagellar tip (Dawson et al., 2007). The dominant-negative overexpression of a rigor mutant kinesin-13 causes significant increases in the length of Giardia's eight flagella, larger median bodies, and cell division defects (Dawson et al., 2007). In contrast, both morpholino-based knockdown of kinesin-2b (Carpenter and Cande, 2009) and dominant negative expression of a rigor mutant kinesin-2a result in significant flagellar shortening and mitotic defects (Hoeng et al., 2008).
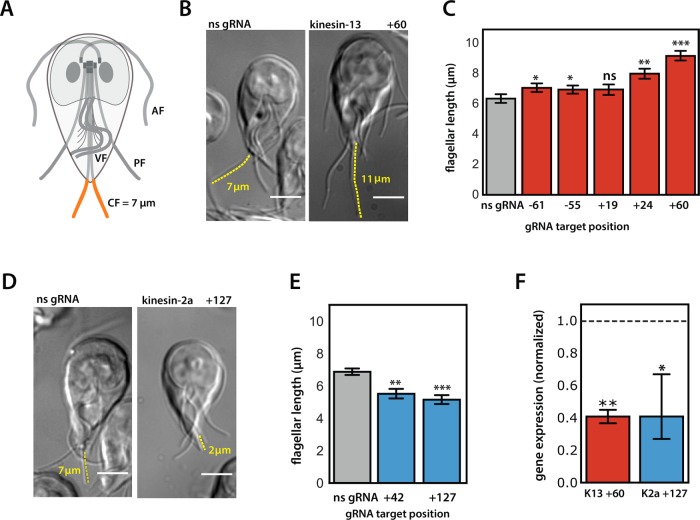
FIGURE 3:
CRISPRi-mediated knockdown of kinesin-13 or kinesin-2a caused significant alterations in flagellar length. Giardia has four pairs of bilaterally symmetric flagella—anterior (AF), ventral (VF), posteriolateral (PF), and caudal (CF, orange)—with distinct equilibrium interphase lengths (A). Representative images and quantification of caudal flagellar length are shown for the CRISPRi-mediated knockdown of two endogenous kinesins known to regulate flagellar length in Giardia: kinesin-13 + 60 gRNA (B) and kinesin-2a +127 gRNA (D) as compared with a nonspecific gRNA strain (see Materials and Methods). Yellow traces highlight representative caudal flagella for flagellar length measurements. Scale bars = 5 µm. In C, average caudal flagellar length for five kinesin-13 knockdown strains with gRNAs targeting the upstream region (–61, –55) or coding region (+19, +24, and +60) are compared with the strain expressing a nonspecific gRNA (ns gRNA). In E, the average caudal flagellar length is quantified for strains with gRNAs targeting +42 and +127 positions of kinesin-2a as compared with a nonspecific gRNA–expressing strain (ns gRNA). All images were acquired from three independent experiments, with more than 70 cells analyzed for each treatment group. In F, the decrease in gene expression of kinesin 13 (red) in the kinesin-13 +60 strain and kinesin-2a (blue) in the kinesin-2a +127 strain is shown relative to expression levels of the two kinesins in a ns gRNA–expressing strain (dotted line indicates expression in the ns gRNA strain). Kinesin gene expression is normalized to the Giardia GAPDH gene from two independent experiments. In all panels, error bars indicate 95% confidence intervals, and significance was assessed using an unpaired t test with ns = not significant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
To determine the ability of CRISPRi to knockdown endogenous genes in Giardia, we designed five gRNAs (–61, –55, +19, +24, +60) to target kinesin-13 (GiardiaDB GL50803_16945) and two gRNAs (+42, +127) to target kinesin-2a (GiardiaDB GL50803_17333). To assess the flagellar length defects of kinesin-13 and kinesin-2a knockdowns, we fixed cells and measured the length of the caudal flagella, a convenient reporter for flagellar length (Dawson et al., 2007). Four of five gRNAs targeting kinesin-13 (–61, –55, +24, +60) significantly increased the length of caudal flagella compared with a nonspecific gRNA (Figure 3, B and C). While both gRNAs targeting the promoter region of kinesin-13 (–61, –55) resulted in significant length increases for caudal flagella, the length defects were less severe than for the two gRNAs (+24, +60) targeting the coding region (Figure 3C). Both gRNAs targeting kinesin-2a (+42, +127) significantly decreased caudal flagellar length compared with that for a nonspecific gRNA (Figure 3, D and E).
To determine the degree of transcriptional repression of kinesin-13 and kinesin-2a conferred by CRISPRi knockdown, we quantified the expression of these targets in the kinesin-13 +60 and kinesin-2a +127 strains using quantitative PCR (qPCR). Expression of either target was reduced by 59% as compared with that of a nonspecific gRNA strain (Figure 3F).
CRISPRi knockdown of MBP causes severe disk defects that are highly prevalent and persist at least 1 wk in cultured trophozoites
Attachment to the host intestinal epithelium by the ventral disk—a highly ordered and complex microtubule (MT) array—is critical to Giardia's pathogenesis in the host (Nosala et al., 2018). The ventral disk is defined by more than 90 parallel and uniformly spaced MTs that spiral into a circular domed structure (Friend, 1966; Holberton, 1973, 1981; Feely et al., 1982; Crossley and Holberton, 1983, 1985). MBP (GiardiaDB GL50803_16343) is one of 87 proteins that localize to the ventral disk (Nosala et al., 2018), and morpholino knockdown of MBP results in ventral disks with an “open” and “flattened” conformation, as well as decreased attachment efficiency (Woessner and Dawson, 2012).
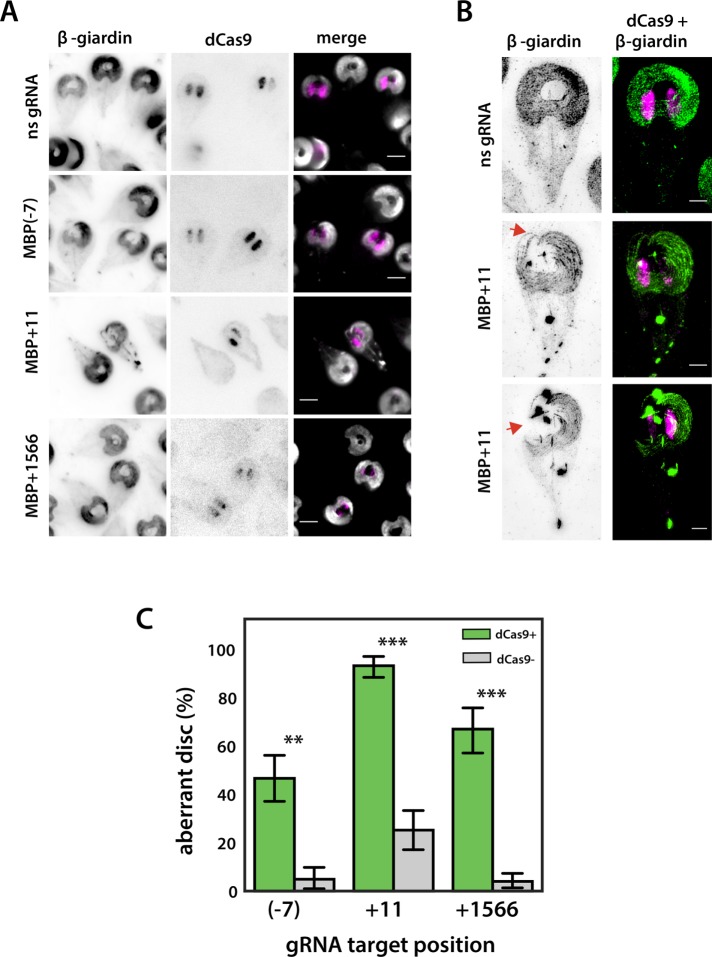
To test the efficacy of MBP knockdown using CRISPRi, we created three strains with gRNAs targeting either the promoter (–7) or coding regions (+11, +1566) of the gene. The proportions of aberrant ventral disk phenotypes in each knockdown strain were determined by fixing and immunostaining trophozoites for dCas9 and the ventral disk protein β-giardin (Baker et al., 1988). Images were scored for disk phenotype and dCas9 expression in each cell (Figure 4A). In each MBP knockdown strain, the presence of an aberrant ventral disk was positively correlated with dCas9 nuclear staining (Figure 4, A and C). Trophozoites positive for dCas9 nuclear staining had severe ventral disk structural defects with incompletely closed (or aberrant) disks (Figure 4, A and B) as compared with the typical closed, domed disk structure (Woessner and Dawson, 2012). Specifically, 46–93% of trophozoites with dCas9 nuclear staining had aberrant ventral disk structures using any of the three gRNAs (Figure 4C). Ventral disks in the nonspecific gRNA strain lacked structural defects and were comparable to wild type. Superresolution imaging using structured illumination microscopy (SIM) highlights the disrupted and open ventral disk ultrastructure in the CRISPRi knockdowns (Figure 4B), with some dCas9-positive cells lacking large regions of the ventral disk (Figure 4B).
FIGURE 4:
CRISPRi knockdown of the ventral disk protein MBP causes severe structural defects. In A, widefield images are presented showing immunostaining of both the disk protein β-giardin (left) and dCas9 (middle) in strains expressing a nonspecific gRNA, or CRISPRi gRNAs designed to knock down the median body protein (MBP(-7) gRNA, MBP+11 gRNA, and MBP+1566 gRNA). Scale bars are 5 µm. In B, representative structured illumination microscopy (SIM) images of the MBP+11 gRNA strain immunostained for β-giardin (left) and dCas9 + b-giardin (right) highlight the characteristic “open” disk conformation (arrows) observed with morpholino knockdown of MBP (Woessner and Dawson, 2012, and see Figure 5B). Scale bar = 2 µm. In C, the proportion of aberrant disk phenotypes in strains expressing MBP(-7), MBP+11, and MBP+1566 gRNAs is quantified and compared in cells scored as dCas9-positive (green) or -negative (gray). Three independent experiments were performed, with more than 200 cells analyzed for each gRNA. Error bars indicate 95% confidence intervals. Significance was assessed using an unpaired t test with **p ≤ 0.01 and ***p ≤ 0.001.
To assess the stability and persistence of CRISPRi knockdown phenotypes, we compared the proportion of trophozoites with aberrant ventral disks obtained via the CRISPRi knockdown to those obtained via morpholino knockdown of MBP over the course of 1 wk. Translational knockdown with morpholinos is currently the only knockdown method widely used in Giardia, yet morpholinos are costly and only transiently repress protein expression (Carpenter and Cande, 2009).
To directly compare the severity, penetrance, and persistence of aberrant disks after MBP knockdown using CRISPRi or morpholinos, we fixed and immunostained trophozoites cultured for 24, 48, 72, and 168 h after introduction of an anti-MBP morpholino and compared them with the constitutively expressed CRISPRi MBP + 11 strain. The proportion of cells with aberrant disks was comparable for the two methods at 24 h. With prolonged passage in culture, however, the penetrance of aberrant disk phenotypes in the morpholino knockdown population decreased rapidly, with significantly fewer aberrant disks present at 48, 72, and 168 h (Figure 5C). At 168 h, no aberrant disk phenotypes were observed in the morpholino knockdown population. In contrast, the penetrance (e.g., the proportion of aberrant disk phenotypes in the CRISPRi strain) remained close to 60% of total cells for 168 h (Figure 5C). Furthermore, more than 85% of dCas9-positive cells had aberrant disk phenotypes at 24 h, and this high degree of penetrance persisted to 168 h of passage in culture (Figure 5D).
FIGURE 5:
Aberrant disk phenotypes in the CRISPRi MBP+11 knockdown strain are highly penetrant and stable. The open-disk phenotype of the MBP+11 gRNA strain (A) is similar to aberrant disk phenotypes observed using an anti-MBP morpholino (B; and see Woessner and Dawson, 2012). DAPI = green, anti-Cas9 = pink. Scale bar = 5 µm. Unlike the anti-MBP morpholino knockdown (orange), the open-disk phenotype of the CRISPRi MBP+11 strain (green) is stable in cultured cells for at least 1 wk (C). The proportion of dCas9+ (green) cells with the open-disk phenotype is also consistent and highly penetrant in the MBP+11 strain (D). Three independent experiments were performed, with more than 75 cells analyzed for each gRNA. Error bars indicate 95% confidence intervals. Significance was assessed using an unpaired t test with ns = not significant, **p ≤ 0.01, ***p ≤ 0.001.
Simultaneous knockdown of kinesin-13 and MBP results in trophozoites with prevalent mutant phenotypes
Simultaneous repression of multiple genes is commonly used to interrogate epistatic interactions or functional redundancy of paralogous genes. The flexibility of the CRISPR/Cas9 system we designed for Giardia permits the simultaneous targeting of multiple regions of the same target or of multiple target genes by simply including multiple gRNAs. To test these versatile aspects of CRISPRi in Giardia, we modified the dCas9g1pac vector to simultaneously express two different gRNAs. In short, we designed a set of primers to amplify the entire gRNA expression cassette from an existing gRNA vector and added this amplified region downstream of the gRNA cassette of a second gRNA vector (Figure 6A). Specifically, we added the MBP + 11 gRNA cassette to the kinesin-13 + 60 vector, as each of these gRNAs provided strong repression of their respective targets with distinct phenotypes (Figures 3C and 4B).
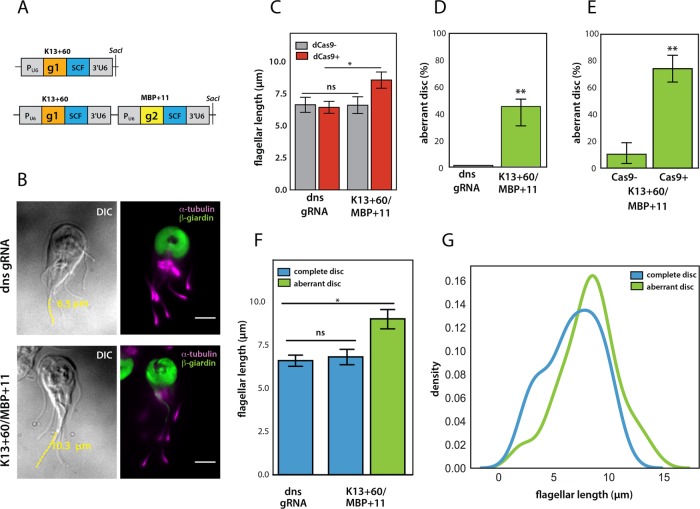
FIGURE 6:
Simultaneous knockdown of kinesin-13 and MBP using two gRNAs to target both genes. For simultaneous expression of two gRNAs targeting both MBP and kinesin-13, we cloned the two gRNA cassettes g1 (MBP + 11) and g2 (kinesin-13 + 60) in tandem. Scaffold sequence = SCF (A). The gRNA cassette is expressed using the Giardia U6 spliceosomal RNA pol III promoter (PU6). In B, representative images for the knockdown of both kinesin-13 and MBP in a strain expressing both kinesin-13 + 60 and MBP + 11 gRNAs highlight a cell with both the open-disk and long-flagella phenotypes as compared with a cell expressing the dual nonspecific gRNAs (dns gRNA). dCas9-positive (red) cells of the K13 + 60/MBP + 11 dual knockdown strain had significantly longer caudal flagella than dCas9-negative cells or those expressing the nonspecific gRNAs (C). Aberrant disk phenotypes (green) were observed in over 45% of cells in the K13 + 60/MBP + 11 dual knockdown strain (D), with high penetrance of aberrant disk phenotypes in dCas9+ cells (E). Cells with aberrant disks (green) also had significantly longer caudal flagella than dns gRNA or K13 + 60/MBP + 11 cells with complete disks (blue) (F). Shifts in the caudal flagellar length distributions (represented using kernel density estimates) show that cells with long flagella have open-disk phenotypes (G). Two independent experiments were performed, with more than 95 cells analyzed for each gRNA. Error bars indicate 95% confidence intervals. Significance was assessed using an unpaired t test with ns = not significant, *p ≤ 0.01 and **p ≤ 0.001.
To assess the efficacy of targeting multiple genes with CRISPRi, we fixed and stained trophozoites expressing dCas9 and the kinesin-13 + 60 and MBP + 11 gRNAs. Flagellar and disk phenotypes were compared with a dCas9 “dual” nonspecific gRNA (dns gRNA) control strain that expressed the nonspecific gRNA sequence from two gRNA cassettes. We quantified flagellar length and the presence of open disks within the same cell and found that a high proportion of the kinesin-13 + 60/MBP + 11 cells had both mutant phenotypes (Figure 6B).
Caudal flagella were 16% longer in the kinesin-13 + 60/MBP + 11 knockdown strain than in the dual nonspecific gRNA strain. As in the single MBP + 11 strain (Figure 3C), trophozoites in the kinesin-13 + 60/MBP + 11 knockdown strain that were positive for dCas9 staining had more severe defects, with caudal flagella that were 30% longer than in cells lacking dCas9 staining (Figure 6C). With respect to ventral disk phenotypes, 45% of kinesin-13 + 60/MBP + 11 knockdown trophozoites had an aberrant “open” ventral disk structure, while no ventral disks in the dual nonspecific gRNA strain had structural defects (Figure 6D). As with caudal flagellar–length defects, open or aberrant ventral disk phenotypes were highly prevalent in dCas9-positive cells; 74% of dCas9-positive cells had aberrant ventral disks as compared with only 10% of dCas9-negative cells (Figure 6E). Last, trophozoites with aberrant disks were primarily found in the fraction of cells with longer caudal flagella (Figure 6F), and especially in cells with caudal flagellar lengths greater than 10 µm (Figure 6G).
DISCUSSION
CRISPRi is a robust alternative to RNAi-mediated gene silencing for precise knockdown of gene expression in both eukaryotic and bacterial model systems (Larson et al., 2013; Kampmann, 2018). Our successful use of CRISPRi to repress both exogenous (Figure 2) and single or multiple endogenous (Figures 3–6) genes in Giardia underscores the versatility of this stable, modular, and efficient gene regulation system. CRISPRi-based gene repression will rapidly change how we study basic cell biology, development, and pathogenesis in this widespread and understudied binucleate parasite.
Development of CRISPRi-mediated knockdown in Giardia
Molecular genetic tool development in Giardia has focused on transient translational repression by electroporation of morpholinos (Carpenter and Cande, 2009) or on the overexpression of long double-stranded RNAs or hammerhead ribozymes for transcriptional repression (Dan et al., 2000; Chen et al., 2007). While CRISPR/Cas9-mediated knockout strategies have recently been used for genome engineering in several parasitic protists (Ren and Gupta, 2017), this is the first demonstration of CRISPRi-mediated transcriptional repression in these organisms.
The Giardia CRISPRi expression system is compact and self-contained on a single episomal plasmid (Figure 1G). Expression of the modular dCas9 and gRNA CRISPRi cassettes does not require Giardia host or viral factors, as are required for antisense (Rivero et al., 2010) or hammerhead ribozyme-mediated transcriptional repression (Dan et al., 2000; Chen et al., 2007). A native Giardia NLS is required for targeting of the Cas9 or dCas9/gRNA DNA recognition complex to both nuclei (Figure 1, E–G). Once imported into the nuclei, the dCas9/gRNA complex is targeted to a specific genomic locus where it sterically interferes with RNA polymerase or transcription factor binding, or with transcriptional elongation, as has been shown in bacteria or other eukaryotes (Larson et al., 2013). The ability to direct dCas9 or other native transcriptional elements to both nuclei in Giardia will enable the modulation of transcriptional networks not only by repression, but also by differential expression through the fusion of dCas9 to Giardia-specific transcription factors (Kampmann, 2018).
The choice of gRNA target site impacts the degree of transcriptional repression in every CRISPRi system (Larson et al., 2013). In both bacteria and human cell lines, targeting gRNAs close to the translation start site results in stronger repression (Gilbert et al., 2013; Qi et al., 2013). We systematically targeted eight sites within the coding region of exogenously expressed NanoLuc (NLuc), a luminescent reporter gene, to determine how gRNA positioning might influence the magnitude of transcriptional repression in Giardia (Figure 2).
Guide RNAs targeting all regions of the NanoLuc reporter substantially repressed luminescence; however, there was no correlation between gRNA target site position and the magnitude of repression. Because we used a native Giardia promoter (MDH) for NanoLuc expression, we could not target this region without potentially altering expression of the native MDH gene. This limits our interpretation of knockdown results for the gRNA that targeted the region immediately upstream of NanoLuc. For the kinesin-13 and MBP CRISPRi knockdowns, however, significant transcriptional repression and aberrant phenotypes were observed for gRNAs targeting regions both upstream and downstream of the translation start site (Figures 3 and 4). More robust and persistent phenotypes for kinesin-2a, kinesin-13, and MBP were associated with gRNAs that targeted the coding regions of these genes, which is consistent with the inhibition of transcriptional elongation, rather than inhibition of transcriptional initiation (Larson et al., 2013). Furthermore, due to the ill-defined nature of Giardia promoters and the limited length of intergenic regions (Davis-Hayman and Nash, 2002), we recommend that several gRNAs be designed to target the coding region for successful repression in Giardia.
Highly penetrant and persistent transcriptional repression of endogenous genes
Using the eight flagella, motile Giardia trophozoites colonize the upper gastrointestinal tract (Dawson and House, 2010), attaching extracellularly to the intestinal villi using the ventral disk, thereby resisting peristaltic flow (Elmendorf et al., 2003; Nosala and Dawson, 2015). Our demonstration of efficient and highly penetrant CRISPRi-mediated transcriptional repression of three endogenous cytoskeletal proteins (Figures 3–6) highlights the versatility of this method of interrogating the contribution of the cytoskeleton to parasite motility and attachment.
Each of the CRISPRi-mediated knockdowns of endogenous genes resulted in cytoskeletal phenotypes that were consistent with phenotypes observed in Giardia in prior studies (Carpenter and Cande, 2009; Dawson et al., 2007; Hoeng et al., 2008; Woessner and Dawson, 2012). In Giardia and other flagellates, alterations in flagellar assembly or disassembly dynamics cause flagellar length defects (Carpenter and Cande, 2009; Dawson et al., 2007; Hoeng et al., 2008; Sloboda, 2005; Woessner and Dawson, 2012). Both electroporation of anti–kinesin-2b morpholinos (Carpenter and Cande, 2009) and inducible dominant negative overexpression of kinesin-2a (Hoeng et al., 2008) limit the degree of IFT-mediated assembly, resulting in shorter flagella in Giardia. In contrast, overexpression of dominant negative kinesin-13 results in increased length of all eight flagella by decreasing the rate of disassembly (Dawson et al., 2007; Hoeng et al., 2008). When we knocked down these kinesins using CRISPRi with gRNAs targeting upstream or downstream of the translation start site, we observed flagellar length increases (up to 60% longer for kinesin-13; Figure 3, B and C) or decreases (up to 30% shorter for kinesin-2a; Figure 3, D and E). These length variations are comparable to those seen with morpholino knockdowns or overexpression of dominant negatives for kinesin-13 or kinesin-2a (Dawson et al., 2007; Hoeng et al., 2008), underscoring the evolutionarily conserved and essential roles these two kinesins play in flagellar length regulation in Giardia.
Giardia's ventral disk is essential for attachment to the host, and is composed of nearly 90 proteins (Nosala et al., 2018). The CRISPRi-mediated knockdown of the disk-associated protein median body protein (MBP) also confirms the same “open” and “flat” disk phenotype (Figure 4) observed using transient morpholino translational knockdowns (Woessner and Dawson, 2012; Figure 5, A and B). Similarly to prior morpholino knockdowns, we also saw extreme defects in ventral disk structure, including partial disks that were most evident with the gRNA targeting the +11 position of the MBP coding region (Figure 4B).
The overall penetrance and stability of CRISPRi knockdown mutant phenotypes in a population are critical for evaluating the efficacy of the Giardia CRISPRi knockdown system. For CRISPRi MBP knockdown with any of three gRNAs, the degree of dCas9 expression was highly correlated with aberrant disk structure (Figure 4C). The penetrance of aberrant disk phenotypes was 46–93% in the population of dCas9-positive cells as compared with 4–25% penetrance in dCas9-negative cells. Thus, in addition to testing multiple candidate gRNAs for severity of phenotypes, we also advocate the use of FACS or similar methods of cell sorting to enrich for a more homogeneous population expressing dCas9, as has been done in other CRISPRi systems (Gilbert et al., 2013).
The stability or persistence of the CRISPRi knockdown phenotype in a population over many generations is also a key feature of the Giardia CRISPRi system, which includes a positive selectable marker (pac) to allow maintenance of the CRISPRi plasmid under puromycin selection. By passaging the MPB + 11 strain repeatedly for 1 wk, we showed that the penetrance of the aberrant disk phenotype remained close to 60% of total cells for up to 168 h (Figure 5C). Between 80 and 97% of dCas9-positive cells had open, incomplete ventral disks at any time point up to 168 h in culture (Figure 5D). Thus, CRISPRi knockdowns are not only highly penetrant but also highly stable as compared with transient knockdowns with morpholinos, in which the prevalence of cells with aberrant disks rapidly decreased in the population after 48 h and was completely lost by 168 h (Figure 5).
As compared with transient and costly morpholino knockdowns, CRISPRi produces stable knockdown strains that are positively selected and can be archived and evaluated at any time. The degree of morpholino knockdown depends on the initial transformation efficiency, and morpholino knockdowns require analysis immediately after electroporation, which could introduce phenotypic artefacts or variability. In contrast, multiple CRISPRi knockdown strains can be made using one or more gRNAs that target the same gene, permitting a comparison of mutant phenotypes in different knockdown strains (Figures 2–5). Furthermore, the generation of CRISPRi knockdown vectors can be multiplexed and requires only the purchase and annealing of two short oligomers.
Knockdown of multiple genes by the simultaneous expression of multiple gRNAs
The modular design of the Giardia gRNA expression cassette allows concatenation of two or more gRNAs to target multiple sites on a single gene, or the targeting of more than one Giardia gene for phenotypic analysis (Figure 6). Simultaneous CRISPRi knockdown of both the flagellar length regulator kinesin-13 and the disk-associated protein MBP resulted in highly penetrant flagellar length and disk defects (Figure 6). Furthermore, the majority of cells with flagellar length defects also had aberrant disks (Figure 6F), and the cells with the longest flagella were exclusively those with open, incomplete disks. Thus, the ability to create stable CRISPRi knockdown strains with defects in multiple genes now allows the evaluation of the functional redundancy of similar and paralogous genes in Giardia, as well as the interrogation of epistatic interactions of genes in a biochemical pathway.
Using CRISPRi to identify and evaluate genes critical in giardiasis
More than 40% of Giardia's 6000 genes encode “hypothetical” proteins that lack similarity to proteins in the human host (Morrison et al., 2007). Many hypothetical proteins are highly expressed during in vivo infections, yet lack any known cellular function (Pham et al., 2017). Rapid and stable CRISPRi knockdown enables the functional evaluation of such hypothetical genes as druggable targets for giardiasis. Combining stable CRISPRi knockdowns with our recently developed bioluminescent imaging (BLI) to monitor temporal and spatial patterns of Giardia infection dynamics and metabolism in the host (Barash et al., 2017) will allow the examination of Giardia mutants with respect to in vivo fitness, colonization, or encystation defects.
The lack of forward genetic tools for Giardia has limited our ability to define genes that are required for basic parasite biology. CRISPRi knockdowns with partial transcriptional repression facilitate the identification of genes with severe fitness costs, as the complete knockdown or knockout of essential genes results in lethal phenotypes. In contrast to reverse genetic approaches, the use of untargeted, genome-wide CRISPRi screens (Larson et al., 2013; Kampmann, 2018) could identify essential genes critical for Giardia growth, differentiation, and pathogenesis.
MATERIALS AND METHODS
Construction of the expression vector Cas9-SV40NLS-GFP for Cas9 localization
The expression vector Cas9-SV40NLS-GFP was constructed by placing mammalian codon–optimized S. pyogenes Cas9 (mCas9) with a C-terminal SV40 nuclear localizing signal (NLS) from JDS246 (Addgene #43861; J.K. Joung, Massachusetts General Hospital, Charleston, MA, and Harvard Medical School, Boston, MA, unpublished data) under the control of the Giardia malate dehydrogenase (MDH; GiardiaDB GL50803_3331) promoter. A 3xHA epitope tag widely used in Giardia (Gourguechon and Cande, 2011) was added to the C-terminal end and the resulting PMDH-mCas9-SV40NLS-3HA fragment was cloned into our C-terminal GFP vector, pcGFP1Fpac (Hagen et al., 2011), fusing Cas9 to GFP.
Identification of putative Giardia NLSs and construction of vectors to test NLS efficacy
Putative NLS sequences were identified among the protein sequences of 65 Giardia strains expressing C-terminal GFP-tagged proteins that localize to the nuclei (Aurrecoechea et al., 2009; Hagen et al., 2011) using the NLS prediction software NLStradamus (Nguyen Ba et al., 2009), with the two-state HMM static model and posterior prediction with a cutoff of 0.6. To delete C-terminal NLSs, we amplified protein-coding regions plus 200 base pairs of upstream sequence to include native promoters using reverse primers that excluded the NLSs and any downstream sequence. The truncated fragments were cloned into pcGFP1Fpac (Hagen et al., 2011). NLS sequences were added to Cas9 either by replacing the GFP tag in Cas9-SV40NLS-GFP with the NLS sequence to be tested (for C-terminal NLSs) or by adding a new start codon and NLS in front of the start codon of Cas9 (for N-terminal NLSs). NLS sequences were generated by PCR amplification from genomic DNA or by annealing short complementary oligonucleotides.
Construction of dCas9 CRISPR interference vector dCas9g1pac
dCas9g1pac was created by amplifying the GL50803_2340 NLS from Giardia genomic DNA with 2340NLSAgeF (5′-actgctaccggtcctcccagagaagaagcggtccaag-3′) and 2340NLSNotIR (5′-actgctgcggccgctttagctcttaattttactaactctacgatcc-3′) and cloning it into Cas9-SV40NLS-GFP to replace GFP. A gRNA expression cassette with inverted BbsI sites for cloning specific gRNA targeting sequences was added, followed by the gRNA scaffold sequence from pX330 (Cong et al., 2013). To drive gRNA expression in Giardia and ensure proper termination, we included ∼200 base pairs of DNA located upstream and downstream of the Giardia U6 spliceosomal snRNA (Hudson et al., 2012). The upstream sequence includes 12 base pairs of U6 coding sequence, and the downstream sequence includes the 12–base pair motif thought to mediate 3′ end processing (Hudson et al., 2012). The gRNA cassette was synthesized (Biomatik) and inserted into a unique Acc65I site in the vector. The dCas9 mutations D10A and H840A were added by replacing a 3.6-kb NcoI–NheI fragment within Cas9 with a similar fragment from dCas9 vector MSP712 (Addgene #65768; Kleinstiver et al., 2015). As constructed, the dCas9g1pac vector contains an 18–base pair nonspecific sequence between the U6 promoter and gRNA scaffold sequence that also includes the inverted BbsI sites for cloning annealed guide oligos. All BLAST hits for this sequence to the Giardia ATCC 50803 genome lack the protospacer adjacent motif (PAM) that is essential for CRISPRi silencing (Larson et al., 2013); all also have mismatches in the critical “seed region” required for dCas9 binding (Qi et al., 2013) or have six or more mismatches throughout the guide sequence. Trophozoites carrying the “nonspecific gRNA” vector have no discernible phenotype.
Guide RNA design and cloning
Specific guide RNAs (20 nt) were designed with the CRISPR “Design and Analyze Guides” tool from Benchling (https://benchling.com/crispr) using a NGG PAM sequence and the G. lamblia ATCC 50803 genome (GenBank Assembly GCA_000002435.1). gRNAs were designed to target the nontemplate strand unless otherwise specified (Supplemental Material). gRNA oligonucleotides with four-base overhangs complementary to the vector sequence overhangs were annealed and cloned into BbsI-digested dCas9g1pac. To express two gRNAs, each was individually cloned and then a universal primer set (gRNA_multiplex_F: 5′-gtcttataaggtaccgagcttgattgcaatagcaaacag-3′ and gRNA_multiplex_R: 5′-ctatagggcgaattcgagctgagctcggtaccttataagacaacatc-3′) was used to amplify the entire gRNA cassette from one plasmid and insert it into the SacI site of the second via Gibson assembly (Gibson et al., 2009). This method was also used to generate the nonspecific gRNA plasmid dCas9g2pac, which has two gRNA cassettes, each with the nonspecific gRNA sequence used in dCas9g1pac. While we have used only two gRNAs here, this method regenerates the SacI restriction site, allowing the insertion of additional gRNAs using the same primers (Supplemental Material).
Construction of the NanoLucNeo luciferase expression vector
The NanoLucNeo vector was constructed via PCR amplification of the MDH promoter (GiardiaDB GL50803_3331), and NanoLuc (pFN31K, Promega). The resulting amplicons were cloned via Gibson assembly into BamHI- and EcoRI-digested pKS_mNeonGreen-N11_NEO (Hardin et al., 2017).
Strains and culture conditions
All G. lamblia (ATCC 50803) strains were cultured in modified TYI-S-33 medium supplemented with bovine bile and 5% adult and 5% fetal bovine serum (Keister, 1983) in sterile 16-ml screw-capped disposable tubes (BD Falcon) and incubated upright at 37°C without shaking. Vectors were introduced into WBC6 or NanoLucNeo trophozoites by electroporation (∼20 µg DNA) as previously described (Hagen et al., 2011). Strains were maintained with antibiotic selection (50 µg/ml puromycin and/or 600 µg/ml G418) (Hagen et al., 2011). All CRISPRi strains were thawed from frozen stocks and cultured for 24–48 h prior to phenotypic analysis, unless otherwise specified.
In vitro bioluminescence assays of NanoLuc knockdown strains
Giardia trophozoites were grown to confluency at 37°C for 48 h, iced for 15 min to detach trophozoites, and centrifuged at 900 × g for 5 min at 4°C. Trophozoites were resuspended in 1 ml of cold media, and serial dilutions (10–1, 10–2) were made for enumeration using a hemocytometer. To measure luminescence, three 50-µl aliquots of the 10–2 dilution (∼1000 cells) were loaded into a white opaque 96-well assay plate (Corning Costar) and incubated for 30 min at 37°C. Following incubation, 50 µl of Nano-Glo Luciferase assay reagent (Promega), prepared at a 1:50 ratio of substrate to buffer, was added to each well. Luminescence was analyzed on a VictorX3 plate reader warmed to 37°C, using 0.1-s exposures, repeated every 30 s until maximal signal was detected. Experiments were performed using two independent samples with three technical replicates that were averaged over three different luminescence acquisitions and displayed with 95% confidence intervals. To determine the linear dynamic range of detection for the NanoLucNeo strain, we used 10-fold serial dilutions and plotted the luminescence readings versus the number of cells loaded into each well (Supplemental Material).
Quantitation of transcriptional knockdown using quantitative PCR (qPCR)
RNA was extracted using a commercial kit (Zymo Research). RNA quality was assessed by spectrophotometric analysis and electrophoresis prior to double-stranded cDNA synthesis using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR of kinesin-2a (GiardiaDB GL50803_17333) was performed using 17333_qPCR1F 5′ GCCTCAACCAACTACGACGA 3′ and 17333_qPCR1R 5′ TCAGCACATCCATCGGCTTT 3′. Quantitative PCR of kinesin-13 (GiardiaDB GL50803_16945) was performed with 16945_qPCR3F 5′ CCAATACGCTGCAAAGCCTC 3′ and 16945_qPCR3R 5′ AGCCAGTTTGTCCATACGCA 3′. Analyses were performed using SensiFast No-ROX SYBR-green master mix (Bioline) in an MJ Opticon thermal cycler, with an initial 2-min denaturation step at 95°C followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 10 s. The constitutively expressed gene for glyceraldehyde 3-phosphate dehydrogenase (GAPDH, GiardiaDB GL50803_ 6687) was chosen as an internal reference gene and was amplified with gapdh-F 5′ CCCTTCACGGACTGTGAGTA 3′ and gapdh-R 5′ ATCTCCTCGGGCTTCATAGA 3′ (Barash et al., 2017). Ct values were determined using the Opticon Monitor software and statistical analyses were conducted using custom Python scripts. All qPCR experiments were performed using two biologically independent samples with four technical replicates. Data are shown as mean expression relative to the GAPDH control sample with 95% confidence intervals.
Immunostaining of CRISPRi knockdown strains
Giardia trophozoites grown to confluency were harvested (see above), washed twice with 6 ml of cold 1X HBS, and resuspended in 500 µl of 1X HBS. Cells (250 µl) attached to warm coverslips (37°C, 20 min) were fixed in 4% paraformaldehyde, pH 7.4 (37°C, 15 min), washed three times with 2 ml PEM, pH 6.9 (Woessner and Dawson, 2012), incubated in 0.125 M glycine (15 min, 25°C), washed three more times with PEM, and permeabilized with 0.1% Triton X-100 for 10 min. After three additional PEM washes, coverslips were blocked in 2 ml PEMBALG (Woessner and Dawson, 2012) for 30 min and incubated overnight at 4°C with anti-TAT1 (1:250, Sigma), anti-HA (1:500, Sigma), anti–beta-giardin (1:1000; gift of M. Jenkins, U.S.Department of Agriculture), and/or anti-Cas9 (1:1000, Abcam) antibodies. Coverslips were washed three times in PEMBALG and incubated with Alex Fluor 488 goat anti-rabbit and/or Alex Fluor 594 goat anti-mouse antibodies (1:250; Life Technologies) for 2 h at room temperature. Coverslips were then washed three times each with PEMBALG and PEM and mounted in Prolong Gold antifade reagent with 4’,6-diamidino-2-phenylindole (DAPI) (Life Technologies). All imaging experiments were performed with three biologically independent samples.
Comparing mutant phenotype severity, prevalence, and persistence between CRISPRi and morpholino knockdowns
Morpholino knockdown of median body protein (MBP, GiardiaDB GL50803_16343) was performed as previously described, using the same anti-MBP and mispair morpholino sequences (Woessner and Dawson, 2012). Following electroporation, morpholino knockdown cells were cultured for 24, 48, 72, or 168 h. The CRISPRi MBP + 11 and nonspecific gRNA knockdown strains, generated as described above, were cultured from a frozen stock of fully selected trophozoites passaged for 24, 48, 72, or 168 h. At each time point, trophozoites were harvested, fixed, and stained as described above. Three biologically independent samples were analyzed for each time point and knockdown method.
Flagellar pair length measurements and ventral disk phenotype analysis
Serial sections of fixed trophozoites were acquired at 0.2-µm intervals using a Leica DMI 6000 wide-field inverted fluorescence microscope with a PlanApo ×100, 1.40 numerical aperture (NA) oil-immersion objective. For flagellar pair length measurements, DIC images were analyzed in FIJI (Schindelin et al., 2012) using a spline-fit line to trace the flagella from the cell body to the flagellar tip. Flagellar length data are presented as mean caudal flagellar length with 95% confidence intervals. For analysis of ventral disk phenotypes, the proportion of aberrant ventral disks and dCas9-positive nuclei within a field of view was determined from maximum-intensity projections of processed images. Trophozoites with colocalized DAPI and anti-Cas9 immunostaining were evaluated as positive for dCas9 expression, while cells with undetectable colocalization were scored as dCas9-negative. Colocalization and flagellar length measurements were analyzed, and figures were generated using custom Python scripts.
Superresolution images of trophozoites exhibiting phenotypes typical of median body protein (MBP, GL50803_16343) silencing by dCas9 were collected at 0.125-µm intervals on a Nikon N-SIM structured illumination superresolution microscope with a ×100, 1.49 NA objective, 100 EX V-R diffraction grating, and an Andor iXon3 DU-897E EMCCD. Images were reconstructed in the “reconstruct slice” mode and were used only if the reconstruction score was 8. Raw and reconstructed image quality were further assessed using SIMcheck (Ball et al., 2015); only images with adequate scores were used for analysis. Images are displayed as a maximum intensity projection.
Accession numbers
The Giardia CRISPRi and NanoLuc reporter vectors have been deposited in GenBank with the following accession numbers: dCas9g1pac (MH037009), NanoLucNeo (MH037010).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Awards R01AI077571 and R21AI119791-01 to S.C.D. Plasmids JDS246 (Addgene #43861) and MSP712 (Addgene #65768) were gifts from Keith Joung. Plasmid pKS_mNeonGreen-N11_NEO was a gift from Alex Paredez (University of Washington, Seattle). The anti–beta-giardin antibody was a gift of Mark Jenkins (U.S. Department of Agriculture, Agricultural Research Service, Animal Parasitic Diseases Laboratory). We thank the MCB Light Microscopy Imaging Facility, a UC Davis Campus Core Research Facility, for the use of the Nikon N-SIM structured illumination superresolution microscope.
Abbreviations used:
- CRISPRi
CRISPR interference
- dCas9
catalytically inactive Cas9
- gRNA
guide RNA
- MBP
median body protein
- MDH
malate dehydrogenase
- NLS
nuclear localizing signal
- NLuc
NanoLuc luciferase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0605) on October 31, 2018.
REFERENCES
- Adam RD, Dahlstrom EW, Martens CA, Bruno DP, Barbian KD, Ricklefs SM, Hernandez MM, Narla NP, Patel RB, Porcella SF, Nash TE. (2013). Genome sequencing of Giardia lamblia genotypes A2 and B isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and E (WB and Pig). Genome Biol Evol , 2498–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al (2009). GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res , D526–D530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Holberton DV, Marshall J. (1988). Sequence of a giardin subunit cDNA from Giardia lamblia. Nucleic Acids Res , 7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Demmerle J, Kaufmann R, Davis I, Dobbie IM, Schermelleh L. (2015). SIMcheck: a toolbox for successful super-resolution structured illumination microscopy. Sci Rep , 15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash N, Nosala C, Pham JK, McInally SG, Gourguechon S, McCarthy-Sinclair B, Dawson SC. (2017). Giardia colonizes and encysts in high density foci in the murine small intestine. mSphere , e00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat LM, Bloland PB. (1997). Drug resistance among malaria and other parasites. Infect Dis Clin North Am , 969–987. [DOI] [PubMed] [Google Scholar]
- Bartelt LA, Sartor RB. (2015). Advances in understanding Giardia: determinants and mechanisms of chronic sequelae. F1000Prime Rep , 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernander R, Palm JE, Svard SG. (2001). Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol , 55–62. [DOI] [PubMed] [Google Scholar]
- Carpenter ML, Cande WZ. (2009). Using morpholinos for gene knockdown in Giardia intestinalis . Eukaryot Cell , 916–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li J, Zhang X, Liu Q, Yin J, Yao L, Zhao Y, Cao L. (2007). Inhibition of krr1 gene expression in Giardia canis by a virus-mediated hammerhead ribozyme. Vet Parasitol , 14–20. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science , 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley R, Holberton DV. (1983). Characterization of proteins from the cytoskeleton of Giardia lamblia . J Cell Sci , 81–103. [DOI] [PubMed] [Google Scholar]
- Crossley R, Holberton DV. (1985). Assembly of 2.5 nm filaments from giardin, a protein associated with cytoskeletal microtubules in Giardia . J Cell Sci , 205–231. [DOI] [PubMed] [Google Scholar]
- Dan M, Wang AL, Wang CC. (2000). Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme. Mol Microbiol , 447–456. [DOI] [PubMed] [Google Scholar]
- Davis JJ, Olsen GJ. (2010). Modal codon usage: assessing the typical codon usage of a genome. Mol Biol Evol , 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Hayman SR, Nash TE. (2002). Genetic manipulation of Giardia lamblia. Mol Biochem Parasitol , 1–7. [DOI] [PubMed] [Google Scholar]
- Dawson SC, House SA. (2010). Life with eight flagella: flagellar assembly and division in Giardia . Curr Opin Microbiol , 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. (2007). Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell , 2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebneter JA, Heusser SD, Schraner EM, Hehl AB, Faso C. (2016). Cyst-Wall-Protein-1 is fundamental for Golgi-like organelle neogenesis and cyst-wall biosynthesis in Giardia lamblia. Nat Commun , 13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsson E, Ma’ayeh S, Svard SG. (2016). An up-date on Giardia and giardiasis. Curr Opin Microbiol , 47–52. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Dawson SC, McCaffery JM. (2003). The cytoskeleton of Giardia lamblia . Int J Parasitol , 3–28. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Singer SM, Nash TE. (2000). Targeting of proteins to the nuclei of Giardia lamblia. Mol Biochem Parasitol , 315–319. [DOI] [PubMed] [Google Scholar]
- Feely DE, Schollmeyer JV, Erlandsen SL. (1982). Giardia spp.: distribution of contractile proteins in the attachment organelle. Exp Parasitol , 145–154. [DOI] [PubMed] [Google Scholar]
- Franzen O, Jerlstrom-Hultqvist J, Castro E, Sherwood E, Ankarklev J, Reiner DS, Palm D, Andersson JO, Andersson B, Svard SG. (2009). Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog , e1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend DS. (1966). The fine structure of Giardia muris . J Cell Biol , 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods , 343–345. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell , 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourguechon S, Cande WZ. (2011). Rapid tagging and integration of genes in Giardia intestinalis. Eukaryotic Cell , 142–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybek M, Golonko A, Gorska A, Szczepaniak K, Strachecka A, Lass A, Lisowski P. (2018). The CRISPR/Cas9 system sheds new lights on the biology of protozoan parasites. Appl Microbiol Biotechnol , 4629–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen KD, Hirakawa MP, House SA, Schwartz CL, Pham JK, Cipriano MJ, De La Torre MJ, Sek AC, Du G, Forsythe BM, Dawson SC. (2011). Novel structural components of the ventral disc and lateral crest in Giardia intestinalis. PLoS Negl Trop Dis , e1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al (2012). Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol , 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanevik K, Bakken R, Brattbakk HR, Saghaug CS, Langeland N. (2015). Whole genome sequencing of clinical isolates of Giardia lamblia. Clin Microbiol Infect , 192 e191–e193. [DOI] [PubMed] [Google Scholar]
- Hardin WR, Li R, Xu J, Shelton AM, Alas GCM, Minin VN, Paredez AR. (2017). Myosin-independent cytokinesis in Giardia utilizes flagella to coordinate force generation and direct membrane trafficking. Proc Natl Acad Sci USA , E5854–E5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth MF. (2014). Immunological aspects of Giardia infections. Parasite , 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeng JC, Dawson SC, House SA, Sagolla MS, Pham JK, Mancuso JJ, Lowe J, Cande WZ. (2008). High-resolution crystal structure and in vivo function of a kinesin-2 homologue in Giardia intestinalis. Mol Biol Cell , 3124–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberton DV. (1973). Fine structure of the ventral disk apparatus and the mechanism of attachment in the flagellate Giardia muris . J Cell Sci , 11–41. [DOI] [PubMed] [Google Scholar]
- Holberton DV. (1981). Arrangement of subunits microribbons from Giardia . J Cell Sci , 167–185. [DOI] [PubMed] [Google Scholar]
- Horlock-Roberts K, Reaume C, Dayer G, Ouellet C, Cook N, Yee J. (2017). Drug-free approach to study the unusual cell cycle of Giardia intestinalis. mSphere , e00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House SA, Richter DJ, Pham JK, Dawson SC. (2011). Giardia flagellar motility is not directly required to maintain attachment to surfaces. PLoS Pathog , e1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AJ, Moore AN, Elniski D, Joseph J, Yee J, Russell AG. (2012). Evolutionarily divergent spliceosomal snRNAs and a conserved non-coding RNA processing motif in Giardia lamblia. Nucleic Acids Res , 10995–11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell , 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, et al (2017). Combined CRISPRi/a-based chemical genetic screens reveal that Rigosertib is a microtubule-destabilizing agent. Mol Cell , 210–223 e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick KS, Peattie DA. (1990). In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J Cell Sci (Pt 3), 353–360. [DOI] [PubMed] [Google Scholar]
- Kaczmarzyk D, Cengic I, Yao L, Hudson EP. (2018). Diversion of the long-chain acyl-ACP pool in Synechocystis to fatty alcohols through CRISPRi repression of the essential phosphate acyltransferase PlsX. Metab Eng , 59–66. [DOI] [PubMed] [Google Scholar]
- Kampmann M. (2018). CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem Biol , 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keister DB. (1983). Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg , 487–488. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature , 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol , 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtkova J, Paredez AR. (2017). Use of translation blocking morpholinos for gene knockdown in Giardia lamblia. Methods Mol Biol , 123–140. [DOI] [PubMed] [Google Scholar]
- Land KM, Johnson PJ. (1999). Molecular basis of metronidazole resistance in pathogenic bacteria and protozoa. Drug Resist Updat , 289–294. [DOI] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc , 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang JR, Veening JW. (2017). High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol , 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al (2007). Genomic minimalism in the early diverging intestinal parasite Giardia lamblia . Science , 1921–1926. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. (2009). NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics , 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosala C, Dawson SC. (2015). The critical role of the cytoskeleton in the pathogenesis of Giardia. Curr Clin Microbiol Rep , 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosala C, Hagen KD, Dawson SC. (2018). “Disc-o-fever”: getting down with Giardia's groovy microtubule organelle. Trends Cell Biol , 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Assaf ZJ, Sept D, Timofejeva L, Dawson SC, Wang CJ, Cande WZ. (2011). An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc Natl Acad Sci USA , 6151–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JK, Nosala C, Scott EY, Nguyen KF, Hagen KD, Starcevich HN, Dawson SC. (2017). Transcriptomic profiling of high-density Giardia foci encysting in the murine proximal intestine. Front Cell Infect Microbiol , 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J , 578–589. [DOI] [PubMed] [Google Scholar]
- Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. (2008). Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science , 1530–1533. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell , 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Gupta N. (2017). Taming parasites by tailoring them. Front Cell Infect Microbiol , 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero MR, Kulakova L, Touz MC. (2010). Long double-stranded RNA produces specific gene downregulation in Giardia lamblia. J Parasitol , 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savioli L, Smith H, Thompson A. (2006). Giardia and Cryptosporidium join the ‘neglected diseases initiative’. Trends Parasitol , 203–208. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloboda RD. (2005). Intraflagellar transport and the flagellar tip complex. J Cell Biochem , 266–272. [DOI] [PubMed] [Google Scholar]
- Tao W, Lv L, Chen GQ. (2017). Engineering Halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi. Microb Cell Fact , 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upcroft J, Samarawickrema N, Brown D, Upcroft P. (1996). Mechanisms of metronidazole resistance in Giardia and Entamoeba . Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy , 47. [Google Scholar]
- Upcroft P, Upcroft JA. (2001). Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin Microbiol Rev , 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner DJ, Dawson SC. (2012). The Giardia median body protein is a ventral disc protein that is critical for maintaining a domed disc conformation during attachment. Eukaryot Cell , 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu ZQ, Liu C, Zheng YG. (2016). Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production. Biotechnol Lett , 2153–2161. [DOI] [PubMed] [Google Scholar]
- Zuberi A, Misba L, Khan AU. (2017). CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front Cell Infect Microbiol , 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.