FIGURE 1:
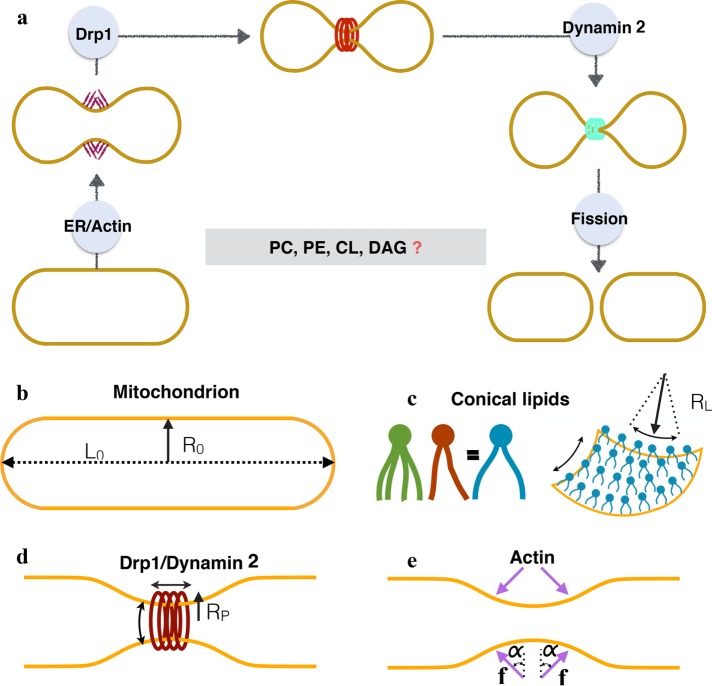
(a) Current working model of mitochondrial fission in mammalian cells. The initial squeezing by actin/ER forces is followed by an increased constriction by a protein Drp1, which is subsequently brought to fission by a protein dynamin 2. While the list of fission proteins has expanded in the recent past, there is also a growing body of evidence that suggests the role of conical lipids in mitochondrial fission (key conical lipids shown in the box). How these lipids collaborate with fission proteins and catalyze fission reaction remains unresolved. In this study we use computational modeling and in vitro experiments to investigate this core puzzle. (b) The basic setup and the key players of membrane remodeling. We simulate an idealized mitochondria in the form of a hollow spherocylinder (a cylinder capped by two hemispheres) made of a lipid bilayer. The geometry is defined by two parameters: the radius R0 and the total length L0. (c) Conical lipids such as PE, CL, and DAG generate spherical curvatures (same in all the directions; see Supplemental Figure S1). Here we suppress the biochemical differences between such lipids and model the effect of a generic cone-shaped lipid with a preferred radius of curvature RL. (d) Drp1 and dynamin 2 are assumed to form a cylindrical coat and generate unequal curvatures in the circumferential and longitudinal directions. The preferred radius of curvature in the circumferential direction RP controls the squeezing capabilities of these proteins. (e) Actin and ER are assumed to apply compressive forces (f) onto the spherocylinder. Because the exact orientation of these forces is not known, we simulate forces at different angles (α).