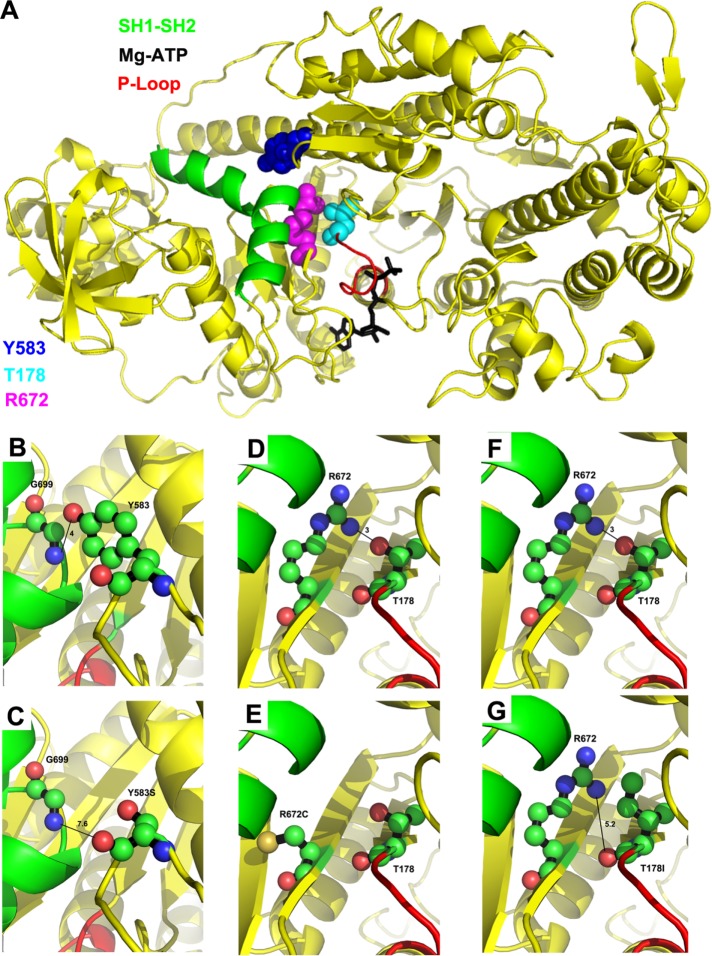
FIGURE 1:
Location of T178, Y583, and R672 on the myosin molecule and interactions disrupted by the FSS mutations. (A) Human myosin FSS residues Y583 (blue), R672 (magenta), and T178 (cyan) in the Drosophila IFM myosin sequence are modeled on to the Dictyostelium discoideum myosin II motor domain structure in the presence of the Mg.ATP complex (PDB #1FMW). Functional domains of interest are illustrated: P-loop (red), SH1–SH2 helix (green), and Mg.ATP complex (black). T178 is located at the N-terminal end of the P-loop. Y583 and R672 are located near the SH1–SH2 helix. (B) Potential hydrogen bonding interaction between Y583–OH and G699–NH, with a contact distance of 4.0 Å. (C) Disruption of potential hydrogen bonding interaction with G699 by the FSS mutation Y583S yielding a new contact distance of 7.6 Å. (D) Potential hydrogen bonding interaction between R672–NH and T178–OH with a contact distance of 3.0 Å. (E) Disruption of potential hydrogen bonding interaction with T178 by the FSS mutation R672C. (F) Potential hydrogen bonding interaction between T178–OH and R672–NH with a contact distance of 3.0 Å. (G) Disruption of potential hydrogen bonding interaction with R672 by the FSS mutation T178I yielding a new contact distance of 5.2 Å. For B–G, carbon, oxygen, and nitrogen atoms are shown in green, salmon, and blue, respectively.