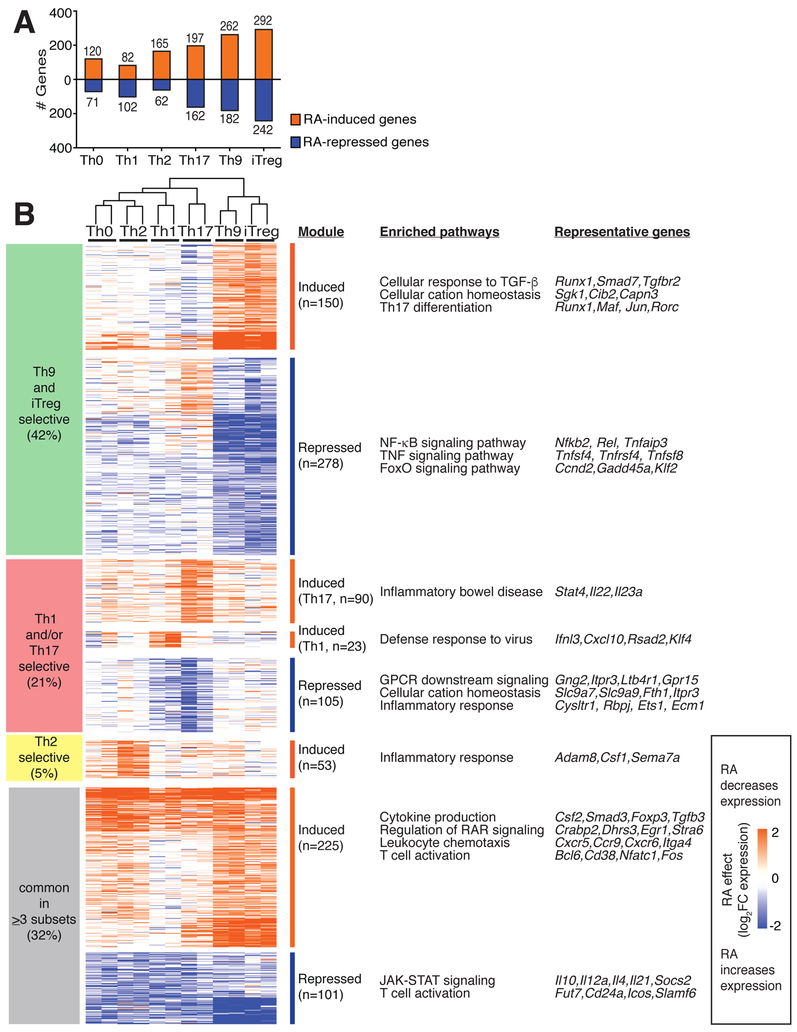
Figure 1: RA has a preferential effect on the transcriptomes of in vitro derived Th9- and iTreg cells.
A. Number of RA-regulated differentially expressed genes (DEGs) in different Th subsets. Isolated naïve CD4+ T cells were cultured with cytokines and antibodies to promote differentiation to the above Th subsets, in the presence of either vehicle control or 1000 nM RA. After 72h, polyadenylated mRNA was isolated and gene expression was measured by RNAsequencing. Total numbers of DEGs are shown for each Th subset (fold-change (FC) in expression with RA vs. vehicle control ≥2 or ≤−2, FDR<0.05). RA regulated 191, 184, 227, 359, 444, and 534 DEGs in Th0, Th1, Th2, Th17, Th9, and iTreg cells. B. RA-regulated genes and pathways. RA regulated a total of 1025 DEGs in ≥1 Th subset. 326 DEGs (32%) were regulated in ≥3 subsets, whereas 699 DEGs (68%) were regulated in ≤2 subsets. 428 DEGs (42%) were primarily regulated in Th9/iTreg cells. Key pathways enriched in each group of DEGs are shown, as well as representative DEGs from each pathway (http://metascape.org, (Tripathi et al., 2015)). (n=2. Heatmap displays log2FC in expression, RA vs. control, see also Figure S1)