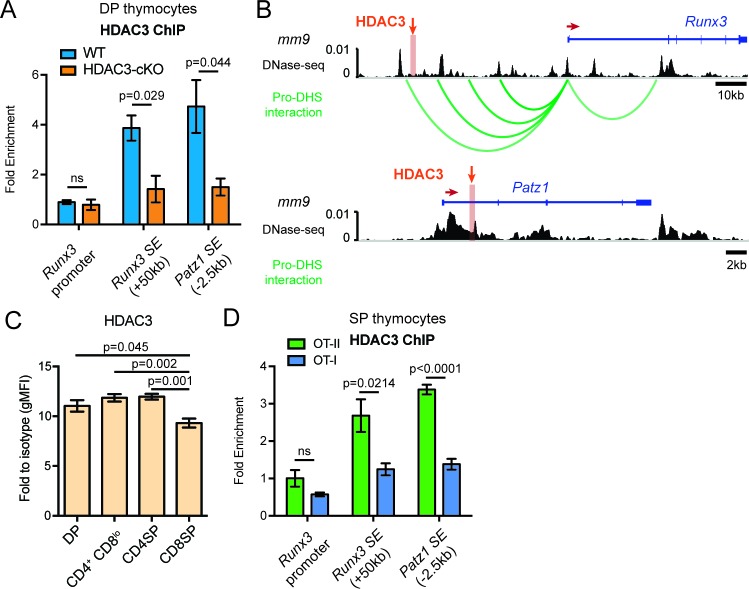
Figure 7. HDAC3 binds to Runx3 and Patz1 regulatory regions in DP and CD4SP thymocytes.
(A) HDAC3 qChIP in DP thymocytes from WT and HDAC3-cKO mice. Plots show mean ± SEM of fold enrichment over Rpl30 (n = 3 mice/group from three independent experiments). (B) DNase-seq and Hi-C arc plots at the Runx3 and Patz1 gene loci in DP thymocytes and pooled DN3-to-DP thymocytes, respectively. Shaded region highlights where HDAC3 binds, as shown in (A). (DHS, DNA hypersensitivity sites). (C) HDAC3 protein expression measured by flow cytometry in DP, CD4+CD8lo, CD4SP, and CD8SP thymocytes from WT mice. Plots show mean ± SEM of fold enrichment over isotype (n = 4 mice/group from three independent experiments). (D) HDAC3 qChIP in SP thymocytes from OT-II and OT-I mice. Plots show mean ± SEM of fold enrichment over Rpl30 (n = 4 mice/group from two independent experiments). See also Figure 7—figure supplements 1–2.
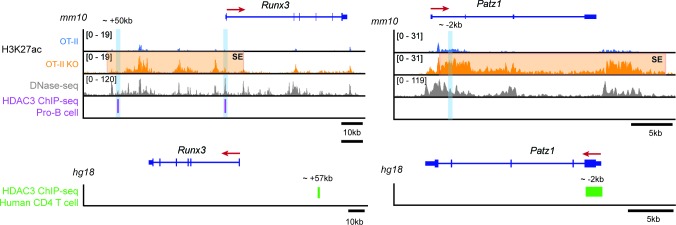
Figure 7—figure supplement 1. HDAC3 ChIP-seq in pro-B cells and human CD4 T cells.
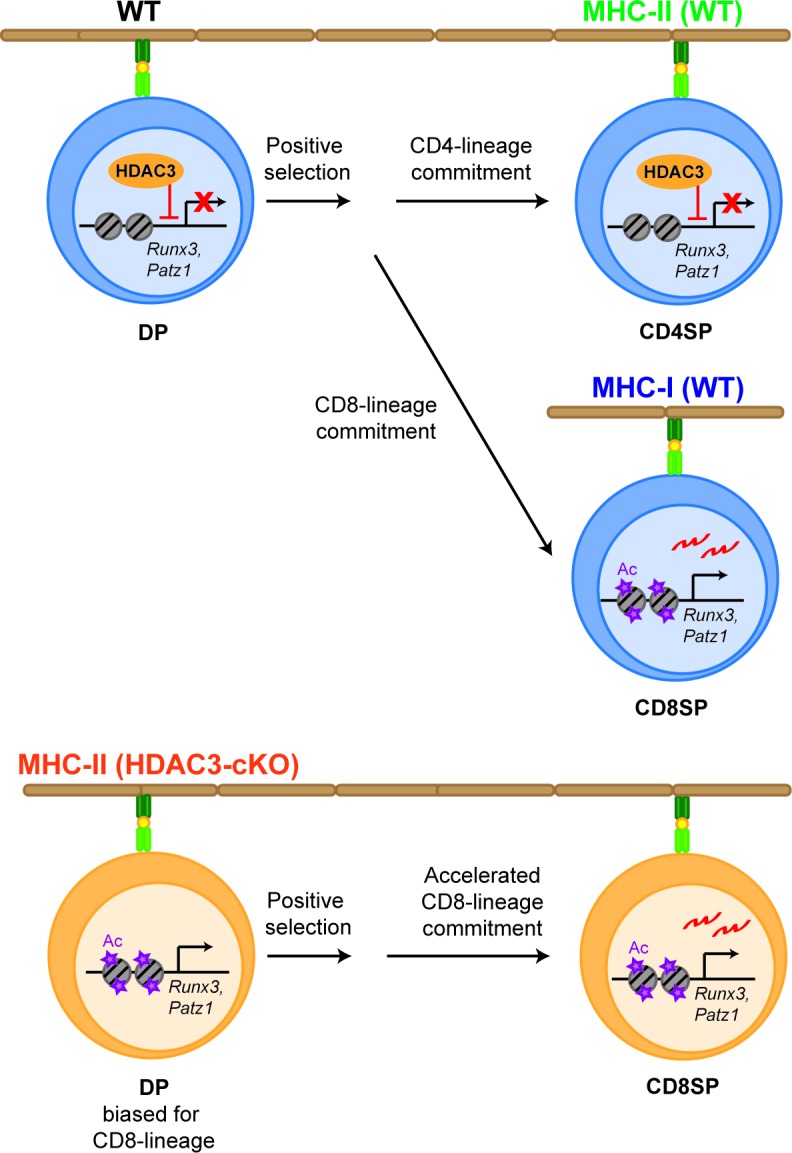
Figure 7—figure supplement 2. Model.