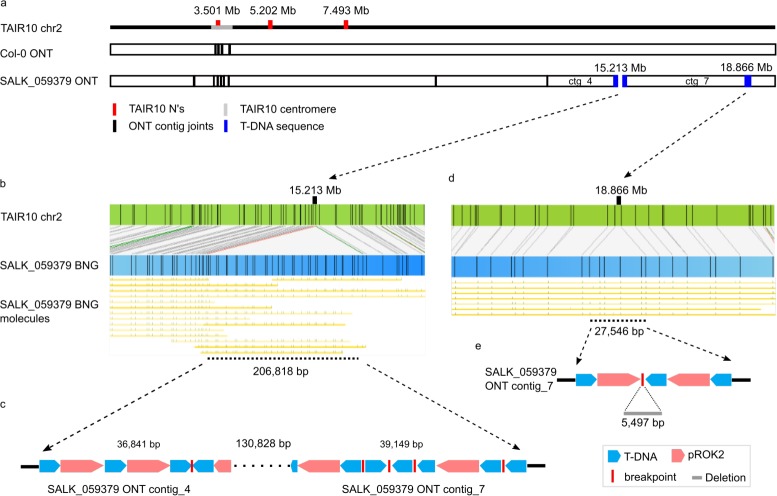
Fig 1. SALK_059379 T-DNA insertions are T-strand and backbone conglomerations.
Schematic representation of Arabidopsis mutant line SALK_059379 chromosome 2 T-DNA insertions as identified from (a, c, e) ONT sequencing de novo genome assemblies and (b, d) BNG optical genome maps. (a) Graphical alignment of Col-0 and SALK_059379 ONT contigs to TAIR10 chromosome 2; two of the three TAIR10 misassemblies (red) are resolved within a single Col-0 ONT contig. Contig joints are represented as vertical black bars. Blue boxes indicate a broken (chr2_15Mb) and an assembled T-DNA insertion (chr2_18Mb). (b, d) represent the BNG maps that identify T-DNA insertions including an alignment of individual BNG molecules. (c) ONT contigs 4 and 7 have the chr2_15Mb insertion site partially assembled and identify a mix of direct T-DNA (blue) and pROK2 backbone (red) concatemers or scrambled stretches with breakpoints as red bars. (e) The chr2_18Mb insertion is assembled and identifies a 5,497 bp chromosomal deletion.