Abstract
Although it has been demonstrated that transformed progenitor cell population can contribute to tumor initiation, factors contributing to this malignant transformation are poorly known. Using in vitro and xenograft based models, previous studies demonstrated that miR-489 acts as a tumor suppressor miRNA by targeting various oncogenic pathways. It has been demonstrated that miR-489 directly targets HER2 and inhibits the HER2 signaling pathway; however, its role in mammary gland development and HER2 induced tumor initiation hasn’t been studied. To dissect the role of miR-489, we sorted different populations of mammary epithelial cells and determined that miR-489 was highly expressed in mammary stem cells. MMTV-miR-489 mice that overexpressed miR-489 in mammary epithelial cells were developed and these mice exhibited an inhibition of mammary gland development in early ages with a specific impact on highly proliferative cells. Double transgenic MMTV-Her2-miR489 mice were then generated to observe how miR-489 overexpression affects HER2 induced tumorigenesis. miR-489 overexpression delayed HER2 induced tumor initiation significantly. Moreover, miR-489 overexpression inhibited tumor growth and lung metastasis. miR-489 overexpression reduced mammary progenitor cell population significantly in preneoplastic mammary glands of MMTV-Her2 mice which showed a putative transformed population in HER2 induced tumorigenesis. The miR-489 overexpression reduced CD49fhiCD61hi populations in tumors that have stem- like properties, and miR-489 overexpression altered the HER2 signaling pathway in mammary tumors. Altogether, these data indicate that the inhibition of HER2 induced tumorigenesis by miR-489 overexpression was due to altering progenitor cell populations while decreasing tumor growth and metastasis via influencing tumor promoting genes DEK and SHP2.
Keywords: miR-489, transgenic mice, mammary gland, HER2/neu tumorigenesis, mammary stem/progenitor cells, breast cancer
Introduction
Breast cancer is a highly heterogenous disease based on phenotypic and functional features. Gene expression profiling of human breast cancer and mouse mammary tumor models broadly classifies breast cancer cells as either luminal- like or basal.12; MMTV-Her2 mouse model is classified as a luminal type breast cancer and MMTV-Her2 mammary tumors have been shown to share gene expression profiles with luminal progenitor cells17. Some of the altered progenitors may function as tumor initiating Cells (TICs), which are responsible for HER2 mediated mammary tumors17, 18, 29. In fact, the cell-of-origin hypothesis suggests that certain breast cancer arise from transformation of stem or progenitor cells30, 31. Therefore, identifying molecular drivers that regulate the stem-progenitor axis may provide insight into the initiation and progression of HER2 mediated tumorigenesis.
Previous studies identified miRNAs as regulators of the mammary stem-progenitor axis and have also been discovered to be dysregulated in breast cancer. For instance, miR-146b, miR-221, miR-199a, miR-182 and miR-193b have been shown to regulate the mammary stem-progenitor axis by targeting various proteins involved in the process3, 9, 14, 19, 33. Also, miR-184 is highly expressed in ducts which proliferate substantially slower than the highly proliferative pubertal terminal end buds, and its expression is lost in mammary tumors of mice. Restoration of miR-184 inhibits proliferation and self-renewal of TNBC cell lines in vivo21.
It has been demonstrated that miR-489 plays a vital role in breast cancer progression and chemo-resistance by targeting several important genes including SMAD3, SPIN1, BCL2 and AKT36, 13. In past reports, we demonstrated that HER2-mediated miR-489 expression via MAPK pathway and negative regulation of HER2 signaling pathway by miR-489 by targeting HER2 and SHP220. Also, miR-489 plays a critical role in muscle stem cell maintenance, where it has been shown to maintain stem cell in a quiescent state by targeting DEK7. Although many studies have reported tumor suppressor roles of miR-489 in breast cancer, its influence on mammary stem cells, mammary gland development, HER2 induced tumor initiation and metastasis remains elusive.
In this study, we dissect the role of miR-489 in the mammary stem-progenitor axis and HER2 induced TICs. To study the role of miR-489 in mammary gland development and HER2 induced tumorigenesis, we developed MMTV-miR489 transgenic mice that specifically overexpress miR-489 in mammary epithelial cells. Using this novel mouse model, we determined the function of miR-489 in progenitor cell regulation. The data show that miR-489 overexpression delayed mammary gland development at early ages and impeded mammary tumor initiation, progression, and metastasis by regulating progenitor cells in the MMTV-Her2 model of breast cancer.
Results and Discussion
miR-489 differentially express in different compartments of mammary epithelial cells
Previously miR-489 was determined to be differentially express in various populations of skeletal muscle with high miR-489 expression in quiescent satellite cells and dramatically lower levels upon entering in to an actively dividing state7. To investigate whether miR-489 has similar functionality in mammary gland, its expression was determined in different sub populations of the mammary epithelial cells. By using florescence activated cell sorting (FACS), purified Lin- mammary epithelial cells from 6-week (wk) old WT mice were separated into four subpopulations: stem-like cells (CD49fhighCD24med) (MRU), myoepithelial cells (CD49fhighCD24low) (Myo), luminal progenitor cells (CD49fmedCD24high) (Ma-CFC) and luminal cells (CD49flowCD24high) (Lum)26, 27 (Fig.1A). Mammary epithelial cells were sorted and characterized by previously demonstrated gene expression analysis25. Our qRT-PCR data demonstrated MRU expressed high level of Keratin14 followed by myoepithelial cells. Since Keratin14 is basal marker, Ma-CFC and luminal cells expressed least amount of Keratin14 (Fig.1B). Luminal cells and Ma-CFC expressed high amount of Keratin18 which is luminal marker (Fig.1C). To further validate MRU population, Krt15, Lgr5 and Lgals1 genes were measured. All three genes were upregulated in MRU as demonstrated previously25 (Fig.1D). miR-489 expression was assayed on each of these populations by qRT-PCR. Higher expression of miR-489 was observed in stem like cells (MRU) compared to Luminal cells, Ma-CFC and myoepithelial cells (MRU vs Lum p=0.0012, MRU vs Myo p=0.0011, MRU vs Ma-CFC p=0.0017) (Fig.1E). miR-489 expression was significantly reduced in Ma-CFC population, which is progenitor cell population (Lum vs Ma-CFC p<0.0001, Myo vs Ma-CFC p=0.0396). This result is consistent with previous in vitro study that showed reduced miR-489 expression in Sca1+ progenitor population of COMMA-Dβgeo cell line compared to Sca1- population11. Together, these data suggest that miR-489 might have regulatory role in stem-progenitor axis. It has been demonstrated that embryonically derived long label retaining cells, which represent quiescent mammary stem cells are restricted to the primary ducts near the nipple region1. Another study demonstrated Lgr5+ mammary epithelial cells, which reside in nipple areas, represent quiescent stem cells22. To determine whether miR-489 expression varies in different parts of mammary gland, RNA was collected from different fractions of inguinal mammary gland of 6-week-old mice as demonstrated in Fig.1F. The results demonstrate that miR-489 is highly abundant in nipple area compared to empty fat pad and terminal end bud (TEB) enriched region (Frac1 vs Frac4 p=0.0004, Frac2 vs Frac4 p=0.0034) (Fig.1G). Observing miR-489 expression in subset of BCL11 or Lgr5+Tspan8hi mammary stem cell populations, which is characterized as quiescent stem cells, will be interesting2, 10. Supporting result is the observation that Lgr5+Tspan8hi population is mostly present in the nipple area. This result might suggest miR-489 may be expressed at higher levels in quiescent mammary stem cells. However, further experiments are warranted to confirm this preliminary observation and to dissect role of miR-489 in maintenance of quiescent mammary stem cell. To find out whether miR-489 is differentially expressed in a temporal manner, RNA was isolated at different stages of mammary gland development including 3.5-wks, 7-wks, 14-wks, day-16 of pregnancy, day-9 of lactation, and involution days-1, 3 and 5. The RT-PCR results clearly demonstrated that miR-489 expression is gradually lost during pregnancy to involution day-3 (Fig.1H). Since pregnancy- activated alveolar progenitor cells are abundant during pregnancy time and miR-489 is least express in progenitor cells28. These data suggest miR-489 expression is higher in mammary stem cells and least in progenitor cells.
Figure 1. miR-489 expression in different sub population of mammary epithelial cells and in the mammary gland during purberty, pregnancy, lactation, and early involution.
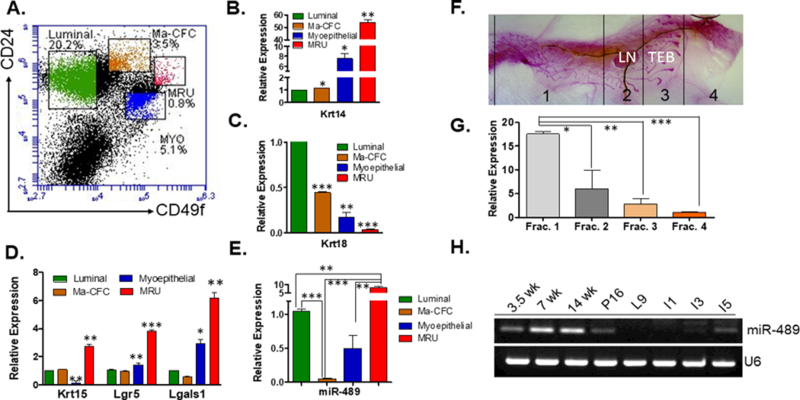
(A) Representative FACS profile for mouse mammary epithelial cells after eliminating hematopoit stained with CD24-PE and CD49f-FITC (B) Keratin14 was assayed by qRT-PCR in luminal, Ma-CFC, myoepthelial cells and MRU. (Lum vs Ma-CFC p=0.0264, Lum vs Myo p=0.0243, Lum vs MRU p=0.0014) (C) Keratin18 was assayed by qRT-PCR in luminal, Ma-CFC, MRU and MRU. (Lum vs Ma-CFC p=0.0003, Lum vs Myo p=0.0039, Lum vs MRU p<0.0001) (D) Krt15, Lgr5 and Lgals1 were measured in different subpopulation of mammary epithelial cells. (For Keratin15 Lum vs MRU p=0.0080 and Myo vs MRU p=0.0056, Lgr5 Lum vs MRU p=0.0008 and Myo vs MRU p=0.0055, Lgals1 Lum vs MRU p=0.0053 and Myo vs MRU p=0.0205) (E) Luminal, Ma-CFC, MRU and myoepithelial subpopulations were collected and assayed for expression of miR-489. (Sorting of cells have been performed 3 independent times and each time 4 sister mice were used, Lum vs MRU p=0.0012, Lum vs Ma-CFC p<0.0001, Myo vs MRU p=0.0011, MRU vs Ma-CFC p=0.0017, Myo vs Ma-CFC p=0.0396) (F) Different fractions of the mammary gland has been collected from 6-week-old WT female as demonstrated. Whole mount image was taken at 4x magnification. (G) RNA was isolated from each fraction and examined for expression of miR-489 (three mice were used for this experiment, Frac1 vs Frac4 p=0.0004, Frac2 vs Frac4 p=0.0034, Frac3 vs Frac4 p=0.0481). (H) RT-PCR data demonstrate miR-489 expression in different stages of mammary gland development. RNA was isolated from three independent mice for each stage. Method: All mice experiments and breeding were performed with approval of the University of South Carolina IACUC. All mice used in this study are on FVB background. For sorting experiment, 6-week-old WT mice were used for this experiment. For each experiment 4 sister mice were used. Thoracic and inguinal mammary glands were used. Glands were minced and digested for 4–6 h at 37°C in DMEM/F12 medium supplemented with 750U/ml Collagenase and 250U/ml hyaluronidase. After this step cells organoids were collected by centrifugation then subjected to ammonium chloride to lyse erythrocytes. After that cells were collected and subjected to separate trypsin (0.5%), dispase (5 mg/ml) and DNase treatment. Cells were passed through a 40µM strainer and subjected for hematopoietic cell depletion. Mouse epithelial cell enrichment kits were obtained from Stem Cell Technologies (catalog#19758) and their protocol was followed. Cells were stained by anti- CD49f (2µl antibody for 1million cells) and anti-CD24 (1.5µl antibody for 1million cells) antibody for 30min on ice. CD49f was obtained from Bio-Legend (cat#313606) and anti-CD24 was obtained from BD bioscience (cat#553262). Different fractions were sorted by using BD FACS aria II sorter. RNA was isolated using Trizole reagent from mammary epithelial cells derived from 3.5, 7 and 14 weeks old nulliparous WT mice. RT-PCR was performed as described in supplementary method to quantitate miR-489 expression with U6 as an internal control20. For rest of the stage female mice were kept with males. For Pregnancy day 16, female mice were kept with male and copulation was confirmed by plug visualization and males removed post coitum. For involution and lactation, pups were removed on day 9 of lactation and mammary glands were collected on day 1, 3 and 5 after the removal of pups. Following primers were used for qRT-PCR; mmu-miR-489–3p 5’-AATGACACCACATATATGGCAGC-3’ and U6 F-5’ GACGTGAGGCGACAAGATGG3’ and R-5’AAAGCCCTTCAGGATACCGC3’. All statistical tests were performed by student t-test using prism (GraphPad software, Inc., San Diego, CA, USA). For qRT-PCR statistical analyses were performed on 2-CT values.
MMTV-miR-489 mice highly express miR-489 in mammary epithelial cells and showed significant delay in mammary gland development at early age
To investigate the role of miR-489 in mammary gland development and tumorigenesis, MMTV-miR-489 mice were generated which only overexpress miR-489 in mammary epithelium (Sup.Fig1A). qRT-PCR data indicate significant up regulation of miR-489 in mammary epithelial cells of MMTV-miR-489 mice. (Sup.Fig.1B) (p<0.0001). Pups were generated by crossing WT male and MMTV-miR-489 female. There was no significant defect on litter weight (Sup.Fig.1C). These data indicate that there is no defect on milk production by miR-489 overexpression. Dek gene has been previously shown to be direct target of miR-489 in muscle tissue7. Furthermore, SHP2 gene has also been identified as a direct target of miR-489 in a variety of human tumor cells15, 20. To test this in mammary tissue, mouse miR-489 mimic was transfected into mouse mammary epithelial cells C127 and 4T1. A significant down regulation of DEK and SHP2 was observed however HER2 expression was not affected by mouse miR-489 overexpression (Sup.Fig1D). To determine if this was extended to the mice, immunoblotting for DEK and SHP2 expression was performed with protein isolated from 6wk mouse mammary epithelial cells from WT and MMTV-miR489 mice. Significant down regulation in DEK and SHP2 expression in MMTV-miR-489 mice were observed (Fig.2A). It is important to find whether Shp2 is direct target gene of miR-489 in mouse system. By searching through 3’UTR sequence of Shp2, we identified a potential complementary binding site (Fig.2B). To demonstrate that this site is functional, we performed the dual luciferase assay using reporter constructs containing 345 bp long wt 3’UTR or mutant 3’UTR of Shp2. Our data showed that in the presence of miR-489, luciferase activity of wt Shp2 and not the mutant Shp2 3’UTR is significantly reduced (Fig.2C). This data demonstrate Shp2 is target of miR-489 in mouse system as well.
Figure 2. Targeted overexpression of miR-489 in mouse mammary epithelial cells impaired mammary gland development in early pubertal age.
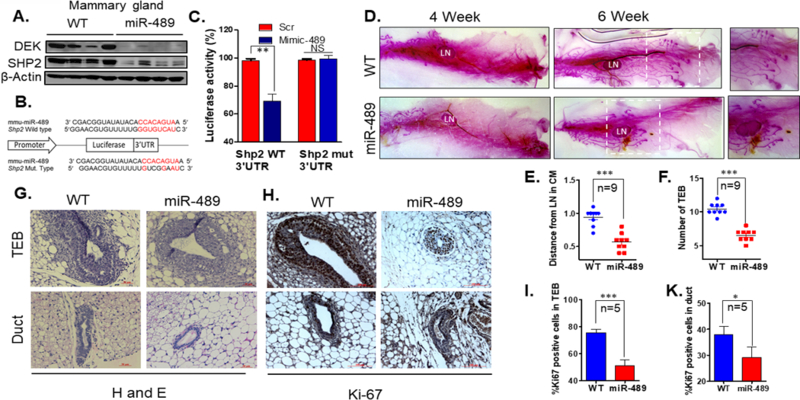
(A) Western blot analysis was performed on protein lysate isolated from mammary epithelial cells from WT and MMTV-miR-489 mice and analyzed for DEK, SHP2 and beta-actin. (n=4 pair of female mice were used) (B) A schematic representation of the Shp2 mRNA with putative miR-489 binding site in the 3’UTR, where seed region is highlighted. (C) H605 cells were co-transfected with either of these vectors with Scramble or miR-489 mimic and renilla expressing vector for 48 h. Firefly luciferase was measured for each condition and normalized with renilla luciferase. (p=0.0096) (D) Whole-mount carmine staining of mammary glands from 4 week and 6 week old virgin mice. Representative images from a sister pair are shown. Whole mount images were taken at 4X magnification. (E, F) Number of Terminal End Bud and distance of TEB from lymph node were measured for analysis purpose (n=9 pair of female mice were used, for number of TEBs p<0.0001 and for distance p<0.0001). (G) Hematoxylin and eosin (H&E) stained representative image of WT and MMTV-miR-489 mouse mammary gland. (H, I, K) Ki-67 stained TEB, duct and histogram of WT and MMTV-miR-489 mammary glands have been demonstrated. (n=5 pair of female mice were used; TEB p<0.0001 and Duct p=0.0441). Method: The full-length mouse pre miR-489 cDNA was amplified by PCR and inserted into a pMMTV vector (a kind gift from Dr. Jeffrey Rosen at Baylor College of Medicine) to generate the plasmid MMTV-miR-489. Pronuclear microinjection to generate transgenic mice was performed by the core facility at MUSC8. Transgenic progeny was identified by southern blot analysis and PCR also was performed with genomic DNA isolated from tail biopsy. The following primer pairs were used to perform genotyping: F-5’ACCAGGTCCTGTTGTAGGTTC3’ and R-5’TACTCTGAGTCCAAACCGGG3’. RNA and proteins were isolated from mammary epithelial cells from WT and MMTV-miR-489 mice. For qRT-PCR analysis 9 pair of sister mice were used and performed as described above. Western blot was performed with a total of 4 pairs of sister mice. DEK and SHP2 antibodies were obtained from Proteintech. Dissected 4th inguinal mammary glands were spread on glass slides and exposed to kahle’s fixative overnight. Glands were stained overnight with carmine alum. Adipose was removed by exposure to xylene overnight. All whole mount images were taken by using Stemi 305 dissecting microscope by using 4X magnification. For whole mount analysis, a total of 9 pairs of sister mice were used. H&E and Ki-67 staining was performed as described in supplementary method. For Ki-67 positive cell counting, 3 independent ducts and TEBs were counted from each mammary gland tissue. Total of 5 pairs of sister mice were used.
It became critical to determine how these molecular changes impacted on mammary gland developmental growth. We performed whole mount by carmine staining and Hematoxylin/eosin (H&E) staining of formalin fixed inguinal mammary gland collected from WT and MMTV-miR-489 mice at 4-wk, 6-wk, 8-wk, involution day-2 (ID2) and involution day-4 (ID4). Whole mount staining of the mammary gland revealed a significant delay in ductal growth and terminal end bud formation at 4-wks and 6-wks between WT and MMTV-miR-489 female mice (Fig.2D-G) (number of TEBs p<0.0001 and p<0.0001). A lesser number of Ki-67 positive cells present in TEB and ducts of miR-489 mice suggests that miR-489 overexpression affected actively proliferating cells (Fig.2H-K) (TEB p<0.0001, Duct p=0.0441). Therefore, there is significant growth difference between WT and transgenic mice. However, miR-489 overexpressing mammary gland captured growth equivalent to WT at 8-wk (Sup.Fig2A). Similarly, mammary glands from ID2 and ID4 had no visible differences but the involution process was observed to occur a little faster in MMTV-miR-489 mice as compared to WT (Sup.Fig2B-D). No significant difference was detected in cleaved caspase-3 positive cells in miR-489 mammary gland at ID4 (Sup.Fig2E). Further study is warranted to confirm the role of miR-489 in the involution process. Together these data demonstrate that miR-489 expression is higher in mammary epithelial cells of MMTV-miR-489 mice and down regulates validated targets in mammary epithelial cells. Further, miR-489 overexpression inhibited mammary gland development by reducing highly proliferative cells in early age.
miR-489 overexpression impedes mammary tumor formation, progression and pulmonary metastasis in MMTV-Her2 mouse model of breast caner
To study the role of miR-489 in Her2 induced spontaneous mammary tumorigenesis in vivo, MMTV-Her2-miR489 mice were generated by intercrossing MMTV-Her2 mice with MMTV-miR-489 mice. These mice overexpress both Her2 and miR-489 in mammary epithelial cells. All mice in this tumor study were nulliparous. MMTV-Her2 transgenic mice have been shown on WT background to develop palpable mammary tumors around 6 months of age and eventually show 100% tumor penetrance. Mammary tumors from MMTV-Her2-miR489 mice successfully overexpresses miR-489 compared to tumors from MMTV-Her2 mice (Sup.Fig3A) (p=0.0016). In MMTV-Her2 mice, palpable mammary tumors appeared as early as 167 days of age with a median onset at 259 days; by 400 days of age all females had palpable tumors as expected (n=28; Fig.3A). In contrast, MMTV-Her2-miR489 mice developed palpable tumors starting at 203 days, with 50% of the mice showing tumors by 379 days and 13% mice didn’t develop any tumor until 570 days (n=23; Fig.3A). Thus, overexpression of miR-489 significantly delayed onset of HER2 induced mammary tumors (P<0.0001). Consistent with this observation, upon sacrificing mice at 8-wk after first palpation of the tumor, we found significant reduction in tumor multiplicity in mice overexpressing miR-489 (Fig.3B) (p=0.0008). Tumor size was measured for the first tumor in each mouse every 7 days after onset. Data showed that the tumors of double transgenic mice were significantly smaller than MMTV-Her2 mice (Fig.3C, Sup.Fig3B) (P=0.0086). Tumor sections from both groups were examined for Ki-67 positive cells, a marker for proliferation. Significantly less number of Ki-67 positive cells were observed in double transgenic mice (P=0.0004; Fig 3D). These data partially explain the significant difference in tumor size between the two groups. To further dissect the underlying mechanism, protein samples isolated from mammary tumors from both groups were analyzed by immunoblotting. There was a significant reduction in SHP2, DEK, pAKT and pERK in MMTV-Her2/miR489 mice compared to MMTV-Her2 mice (Sup.Fig3C-E). However, there was not any difference in HER2 expression upon miR-489 overexpression. One possible explanation for this observation is mouse/rat HER2 is not target of miR-489. But, there is possibility other members of HER2 signaling pathway might affected by miR-489 overexpression. Because of that we observed significant inhibition in HER2 downstream pathway. It has previously been demonstrated that Dek plays an oncogenic role through the paracrine Wnt signaling pathway in Ron receptor positive breast cancers23. It is interesting to explore the role of DEK in HER2- mediated tumorigenesis. Further Kaplan-Meier plot analyses were performed and patients with high DEK expression in their breast tumors, had worse overall survival as compared to patients with low DEK expression (Sup.Fig.4A). It has been shown that SHP2 positively regulates HER2 induced transformation and proliferation by dephosphorylating an autophosphorylation site which serve as docking site for Ras-GTPase activating protein which enhance duration of GTP-Ras levels34. Furthermore, by microscopic examination of lung we found 43% of MMTV-Her2 and MMTV-Her2/miR-489 mice harbored micro-metastasis in the lung. However, double transgenic mice had three times less metastatic foci compared to Her2 transgenic mice (p=0.0035; Fig.3D). Also, Kaplan-Meier plotter analyses demonstrated that HER2 positive breast cancer patients with low miR-489 expression have worse overall survival compared to patients with high miR-489 expression (P=0.00016, Sup Fig4B). This result supported tumor suppressor role of miR-489 in HER2 positive breast cancer. Together, these data suggest restoration of miR-489 might delay HER2 mediated tumor initiation and inhibited tumor growth and their colonization in lung.
Figure 3. Overexpression of miR-489 impedes tumor initiation, growth and lung metastasis in HER2 induced mammary tumors.
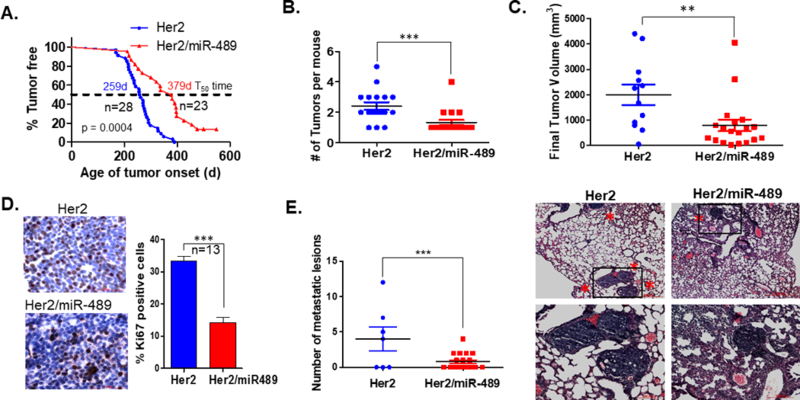
(A) Kaplan-Meier Curves of mammary tumor free survival of virgin MMTV-Her2 (n=15) and MMTV-Her2-miR489 mice (n=20). The p value was calculated using the long rank-rank test (p<0.0001) (B) Mammary tumor multiplicity between MMTV-Her2 (n=17) and MMTV-Her2-miR489 (n=18) mice (P=0.0008) (C) Tumor growth was monitored every 7 days. Final tumor sizes of both group were compared (P=0.0086) (D) Proliferation rate was assessed by Ki-67 immunostaining comparing tumors from MMTV-Her2 (n=10) and MMTV-Her2-miR489 (n=15). At least 500 cells have been counted per mammary tumor. (p<0.0001) (E) Quantitation of the number of metastatic foci in the lungs of MTTV-Her2 and MMTV-Her2-miR489 mice after 8-week post tumor onset. H and E staining of lungs of MMTV-Her2 and MMTV-Her2-miR489 mice. Images were taken at 4x and 10x magnification(n=17) (p=0.0035). Method: MMTV-Neu mice were purchased from Jackson laboratory. MMTV-Neu mice were crossed with MMTV-miR489 mice to generate MMTV-Her2-miR489 double transgenic mice. For analysis of mammary tumor incidence, we monitored adult female mice starting at 4 months of age and once per week. We monitored the mice weekly and noted the time of tumor appearance, tumor size, the number of tumors and lung metastasis in each mouse. After detecting palpable tumor, tumor size was measured every week. Tumor free survival curves were evaluated using Kaplan-meier method and differences between the survival curves were evaluated by a log-rank test. Tumor volume of each primary tumor was calculated by modified ellipsoidal formula (1/2 (LxW2)). Tumor volumes were plotted using graph-pad prism software. For Ki-67 counting 1000 tumor cells were counted from 13 mammary tumors and the percentage of positive staining cells are presented in the figures.
miR-489 overexpression delayed tumor onset by partially affecting progenitor cells which is a major target population for HER2 induced tumorigenesis
Next, we questioned whether miR-489 overexpression has any effect on mammary epithelial cell hierarchy. To address this question, Lin- mammary epithelial cells were obtained from 6wk WT and miR-489 mice. MECs were stained with CD49f and CD24 antibodies to distinguish different population of mammary gland. FACS analysis demonstrates a significant reduction in Ma-CFC population in MMTV-miR-489 mice (p=0.0016; Fig.4A). Similarly, we observed significant reduction of CD49fhiCD24hi population upon overexpression of miR-489 in C127 cells (p=0.002; Sup.Fig5). To validate this observation, a colony formation assay was performed with mammary epithelial cells from WT and miR-489 mice. These data demonstrate a significant decrease in the number of colonies formed using isolated MECs from MMTV-miR-489 mice compared to WT mice (p=0.0038; Fig.4B). Committed progenitor cells are major target for malignant transformation. In our previous study, we observed an increase in progenitor population in MMTV-Her2 mice before tumor onset18. To check whether miR-489 restoration has any effect on Her2 induced progenitor cells, Lin- mammary epithelial cells were analyzed from pre-neoplastic mammary glands from MMTV-Her2 and MMTV-Her2/miR-489 mice. Significant reduction in CD49fmid/CD24high population was observed in Her2/miR489 mice compared to Her2 mice (p=0.0015; Fig.4C). All these results suggest miR-489 restoration might target progenitor cells, which are the major target for malignant transformation by HER2 induced tumorigenesis. These studies have been performed on nulliparous mice. Further study is warranted to observe parity-identified mammary epithelial cells (PI-MEC) which has been also suggested as a target population of HER2/neu-driven tumorigenesis12, 32. Although it has been found that PI-MEC are part of luminal progenitor population4. Previously, we established that CD49fhiCD61hi cells exhibit stem- like traits of self-renewal and regenerating heterogeneity in HER2 induced mammary tumor18. It is important to check whether miR-489 restoration has any effect on this population. We found a significant reduction in CD49fhiCD61hi population by miR-489 expression restoration (p=0.03377; Fig.4D). In summary miR-489 restoration reduces luminal progenitor cell population which are likely targets for malignant transformation in HER2 mediated tumorigenesis.
Figure 4. Overexpression of miR-489 in mammary epithelial cells has reduced Ma-CFC population.
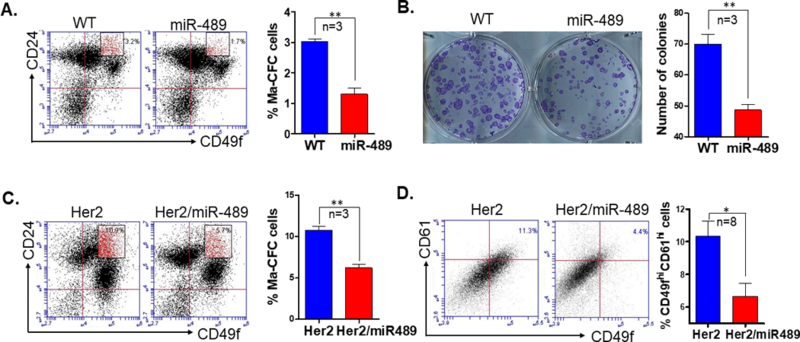
(A) FACS analysis was performed to examine the luminal progenitor cell population (CD49fmedCD24hi) in mammary epithelial cells from 6-week-old WT and MMTV-miR489 transgenic mice (n=3). P=0.0016 (unpaired student t test). (B) Colony formation assay was performed from mammary epithelial cells from 6-week-old WT and MMTV-miR489 transgenic mice (n=3). P=0.0038 (unpaired student t test). (C) Mammary epithelial cells were isolated from 24-week-old Her2 and Her2-miR489 mice and stained with CD49f-FITC and CD24-PE and analyzed for Ma-CFC population (n=3). P=0.0015 (unpaired student t test). (D) FACS analysis of the putative CSC cells (CD49fhiCD61hi population) in the first mammary tumor of each animal (n=8). P=0.0337 (unpaired student t test) Method: Thoracic and inguinal mammary glands were isolated from 6-week-old WT and MMTV-miR-489 mice, single cell mammary epithelial cells suspension were prepared and stained with CD49f and CD24 antibody as described above. Cells were analyzed with BD Accuri C6 flow cytometer for different mammary epithelial cell population. A total of 35,000 events were collected for each sample. Mammary epithelial cells were seeded in plates for Ma-CFC colony formation assay as described in supplementary method. Mammary tumors were harvested, minced and incubated in digestion media to generate single cell suspensions (described above) at 37°C for 2h. Cells were stained with CD49f and CD61 on ice for 30 min. CD61 was obtained from BioLegend (cat#104308).
A growing number of reports have suggested the role of miRNA in breast cancer based on in vitro and xenograft based models. Recently, few miRNA knockout models have been generated to study their roles in autochthonous mammary tumors16, 24. In this study, we demonstrate for the first time the role of miRNA-489 in the mammary stem cell hierarchy and Her2 induced tumorigenesis by using a mammary specific overexpression model in MMTV-Her2 mice. Overexpression of miR-489 reduced progenitor cells, which are the major target population for malignant transformation by HER2 overexpression. This data also suggest miR-489 expression may be one of the factor contributing in malignant transformation of progenitor cells. This can be partially responsible for miR-489 overexpression mediated delay in mammary tumor onset. Further, miR-489 overexpression reduced expression of DEK, SHP2 and HER2 signaling pathways, thus decreasing cell proliferation and inhibiting tumor progression in Her2 induced tumorigenesis model (Sup.Fig6). Recently, we have showed how miR-489 also play tumor suppressor role in triple negative breast cancer by blocking key proteins involve in autophagy5. Our study indicates miR-489 is highly expressed in stem cells and significantly reduced in progenitor cells. Furthermore, miR-489 is at very low expression in TEB which harbors actively dividing stem cells. This may suggest role of miR-489 in quiescent stem cells. We propose a functional model in which miR-489 may be involved in regulation of the quiescent state of stem cells, thereby regulating progenitor cell population. Further studies will be required to validate this hypothesis.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Gene Function and Transgenic and Genome Editing (TGE) Cores of the Medical University of South Carolina (MUSC) for their assistance with the generation of transgenic MMTV-miR-489 mice. This work was supported by NIH grants (4R01 CA178386–04 and 4P30 GM103342–05) and the USC ASPIRE-1 grant to H. Chen, and the USC SPARC graduate research grant to M. Soni.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Boras-Granic K, Dann P, Wysolmerski JJ. Embryonic cells contribute directly to the quiescent stem cell population in the adult mouse mammary gland. Breast cancer research : BCR 2014; 16: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai S, Kalisky T, Sahoo D, Dalerba P, Feng W, Lin Y et al. A Quiescent Bcl11b High Stem Cell Population Is Required for Maintenance of the Mammary Gland. Cell stem cell 2017; 20: 247–260 e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celia-Terrassa T, Liu DD, Choudhury A, Hang X, Wei Y, Zamalloa J et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nature cell biology 2017; 19: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang TH, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast cancer research : BCR 2014; 16: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Soni M, Patel Y, Markoutsa E, Jie C, Liu S et al. Autophagy, Cell Viability and Chemo-resistance are Regulated by miR-489 in Breast Cancer. Mol Cancer Res 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L et al. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. The Journal of pathology 2016; 239: 459–472. [DOI] [PubMed] [Google Scholar]
- 7.Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012; 482: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho A, Haruyama N, Kulkarni AB. Generation of transgenic mice. Curr Protoc Cell Biol 2009; Chapter 19: Unit 19 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsarraj HS, Hong Y, Valdez K, Carletti M, Salah SM, Raimo M et al. A novel role of microRNA146b in promoting mammary alveolar progenitor cell maintenance. Journal of cell science 2013; 126: 2446–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu NY, Rios AC, Pal B, Law CW, Jamieson P, Liu R et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nature cell biology 2017; 19: 164–176. [DOI] [PubMed] [Google Scholar]
- 11.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. Journal of cell science 2010; 123: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C et al. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer cell 2010; 17: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A et al. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS letters 2014; 588: 2009–2015. [DOI] [PubMed] [Google Scholar]
- 14.Ke J, Zhao Z, Hong SH, Bai S, He Z, Malik F et al. Role of microRNA221 in regulating normal mammary epithelial hierarchy and breast cancer stem-like cells. Oncotarget 2015; 6: 3709–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). British journal of cancer 2010; 103: 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Siverly AN, Chen D, Wang M, Yuan Y, Wang Y et al. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer research 2016; 76: 6424–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast cancer research : BCR 2010; 12: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo PK, Kanojia D, Liu X, Singh UP, Berger FG, Wang Q et al. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene 2012; 31: 2614–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Ruiz H, Illa-Bochaca I, Omene C, Hanniford D, Liu Q, Hernando E et al. A TGFbeta-miR-182-BRCA1 axis controls the mammary differentiation hierarchy. Science signaling 2016; 9: ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel Y, Shah N, Lee JS, Markoutsa E, Jie C, Liu S et al. A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK signaling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget 2016; 7: 18295–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phua YW, Nguyen A, Roden DL, Elsworth B, Deng N, Nikolic I et al. MicroRNA profiling of the pubertal mouse mammary gland identifies miR-184 as a candidate breast tumour suppressor gene. Breast cancer research : BCR 2015; 17: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell reports 2013; 3: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Privette Vinnedge LM, Benight NM, Wagh PK, Pease NA, Nashu MA, Serrano-Lopez J et al. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene 2015; 34: 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes & development 2017; 31: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soady KJ, Kendrick H, Gao Q, Tutt A, Zvelebil M, Ordonez LD et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res 2015; 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D et al. Purification and unique properties of mammary epithelial stem cells. Nature 2006; 439: 993–997. [DOI] [PubMed] [Google Scholar]
- 27.Stingl J. Detection and analysis of mammary gland stem cells. The Journal of pathology 2009; 217: 229–241. [DOI] [PubMed] [Google Scholar]
- 28.Tiede BJ, Owens LA, Li F, DeCoste C, Kang Y. A novel mouse model for non-invasive single marker tracking of mammary stem cells in vivo reveals stem cell dynamics throughout pregnancy. PloS one 2009; 4: e8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaillant F, Asselin-Labat ML, Shackleton M, Lindeman GJ, Visvader JE. The emerging picture of the mouse mammary stem cell. Stem cell reviews 2007; 3: 114–123. [DOI] [PubMed] [Google Scholar]
- 30.Visvader JE. Cells of origin in cancer. Nature 2011; 469: 314–322. [DOI] [PubMed] [Google Scholar]
- 31.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell 2012; 10: 717–728. [DOI] [PubMed] [Google Scholar]
- 32.Yallowitz AR, Alexandrova EM, Talos F, Xu S, Marchenko ND, Moll UM. p63 is a prosurvival factor in the adult mammary gland during post-lactational involution, affecting PI-MECs and ErbB2 tumorigenesis. Cell Death Differ 2014; 21: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo KH, Kang K, Feuermann Y, Jang SJ, Robinson GW, Hennighausen L. The STAT5-regulated miR-193b locus restrains mammary stem and progenitor cell activity and alveolar differentiation. Developmental biology 2014; 395: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Agazie YM. Molecular mechanism for SHP2 in promoting HER2-induced signaling and transformation. The Journal of biological chemistry 2009; 284: 12226–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


