Fig. 1.
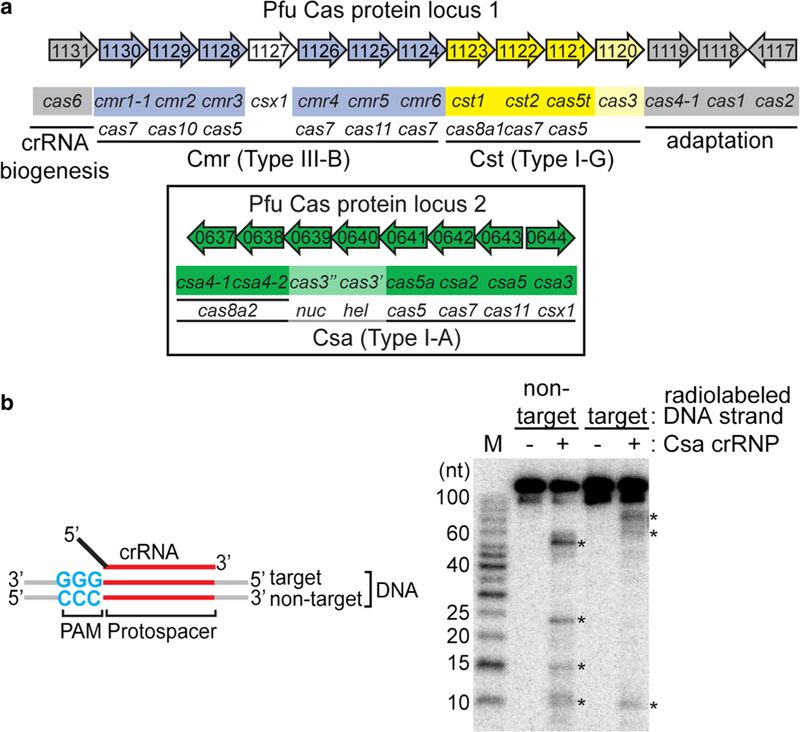
Reconstitution of functional Type I-A crRNPs. a Pyrococcus furiosus (Pfu) cas gene organization and annotation. Genes encoding the crRNA biogenesis protein (cas6, gray), predicted CRISPR DNA integration proteins (cas1, cas2 and cas4, gray) and three effector modules: Type I-A (csa, green and boxed), Type I-G (cst, yellow) and Type III-B (cmr, blue) are shown. The annotated superfamily designation (e.g., cas5, cas7, cas8) is denoted below each gene name. b Schematic of double-stranded DNA substrate used in the assay. Wildtype PAM (blue), protospacer (red), target strand (complementary to crRNA guide) or non-target strand of DNA, 5′ tag (black) and guide (red) of crRNA are labeled. Denaturing PAGE demonstrating cleavage (asterisk) of double-stranded DNA in the absence (−) or presence of Csa crRNP (+) (assembled with six Csa proteins (Csa4–1, Cas3″, Cas3′, Cas5a, Csa2, Csa5) and crRNA). Top label indicates location of 5′ radiolabeling (32P) in either the target strand or non-target strand of double-stranded DNA. 5′ radiolabeled (32P) DNA size-standard (Affymetrix, 10–100 bases) is labeled (M)
