Fig. 5.
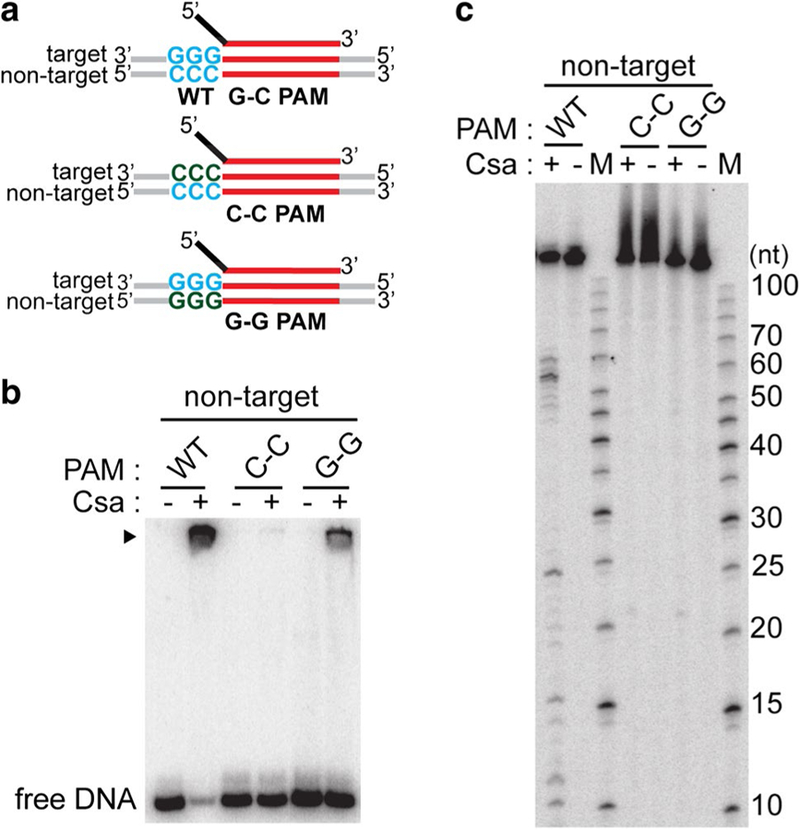
Type I-A crRNPs recognize double-stranded PAM for DNA destruction. a Schematic of double-stranded DNA substrates used in assay: crRNA (top) containing 5′ tag (black) and guide (red), doublestranded DNA (target and non-target strands labeled) with wild-type protospacer (red) and PAM (WT G-C PAM (wild type, blue) or G–G PAM [single-stranded, wild type on target strand (blue) and mutated on non-target strand (Sternberg et al. 2014)] or C–C [single-stranded, wild type on non-target strand (blue) and mutated on target strand (Sternberg et al. 2014)]. b DNA binding and c DNA cleavage with no protein (−) Csa crRNP ((+) assembled with crRNA, Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5) and the specified substrates. DNA bound by Csa crRNP (black triangle) and free DNA in b is indicated. 5′ radiolabeled (32P) DNA size standard (Affymetrix, 10–100 bases) (c) is labeled (M). The non-target strand of DNA is 5′-radiolabeled (32P) in all cases
