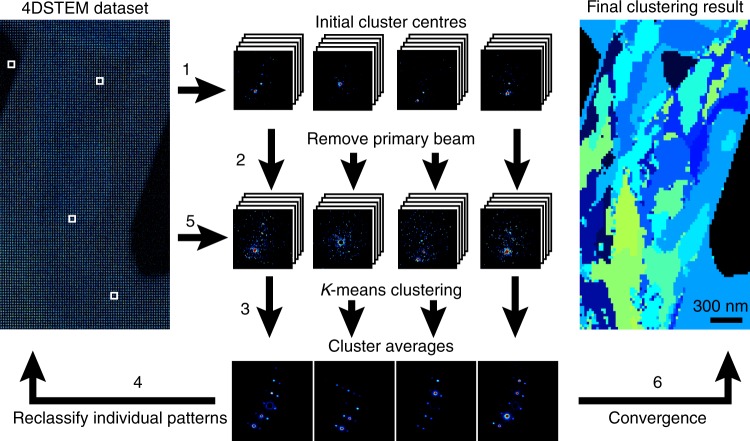
Fig. 2.
Workflow of unsupervised clustering used to define regions of similar diffraction in peptide crystals. (Step 1) Initial cluster 'centres' are assigned within the 4DSTEM dataset (left white boxes) and all patterns are binned. (Step 2) The primary beam is subsequently masked to prevent its influence on clustering. (Step 3) Individual patterns are then compared to each cluster centre sequentially via Euclidean distance and assigned to the cluster where this distance is smallest. (Step 4) Average patterns are calculated for each cluster and (step 5) are used to reclassify individual patterns until convergence (step 6) where a final map illustrating spatial localization of similar diffraction patterns is produced