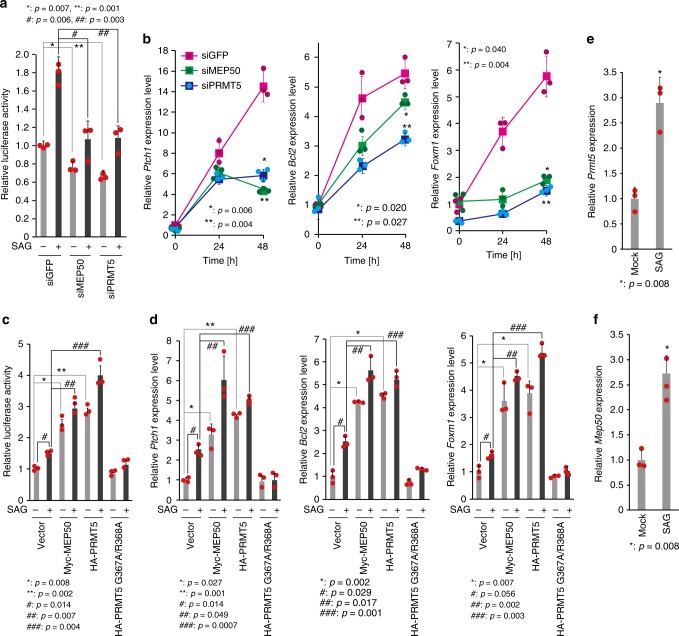
Fig. 3.
MEP50/PRMT5 complex-mediated GLI1 stabilisation enhances Gli transcriptional activity and HH signalling pathway activation induces PRMT5 and MEP50 expression. a Gli transcriptional activity in PRMT5 or MEP50 knockdown cells. siMEP50-m2 and siPRMT5-m2 siRNAs were stably expressed by recombinant retroviruses. A multimerized Gli-binding site luciferase reporter plasmid and phRL-TK control reporter plasmid were transfected into C3H10T1/2 cells. After 24 h of incubation, 300 nM SAG was applied for 24 h, and then luciferase assays were performed. b qRT-PCR analysis of Ptch1, Bcl2, and Foxm1 expression in C3H10T1/2 cells with MEP50 knockdown or PRMT5 knockdown and treated with 300 nM SAG for the indicated times. siMEP50-m2 and siPRMT5-m2 siRNAs were stably expressed by recombinant retroviruses. c Gli transcriptional activity in HA-PRMT5 or Myc-MEP50-expressing cells. HA-PRMT5, HA-PRMT5 G367A/R368A, or Myc-MEP50 and a multimerized Gli-binding site luciferase reporter plasmid and phRL-TK control reporter plasmid were transfected into C3H10T1/2 cells. After 24 h of incubation, 300 nM SAG was applied for 24 h, and then luciferase assays were performed. d qRT-PCR analysis of Ptch1, Bcl2, and Foxm1 expression in HA-PRMT5 or Myc-MEP50-expressing C3H10T1/2 cells. HA-PRMT5, HA-PRMT5 G367A/R368A, or Myc-MEP50 plasmids were transfected into C3H10T1/2 cells. After 24 h of incubation, cells were separated equally, and DMSO (−) or 300 nM SAG (+) were applied for 24 h. Protein levels are shown in Supplementary Fig. 3a. e and f qRT-PCR analysis of PRMT5 (e) and MEP50 (f) mRNA expression in C3H10T1/2 cells after 24 h of treatment with 300 nM SAG. In a–c, data represent one of two independent experiments with similar results. In e and f, data represent one of three independent experiments with similar results. The source data is shown in Supplementary Data 1