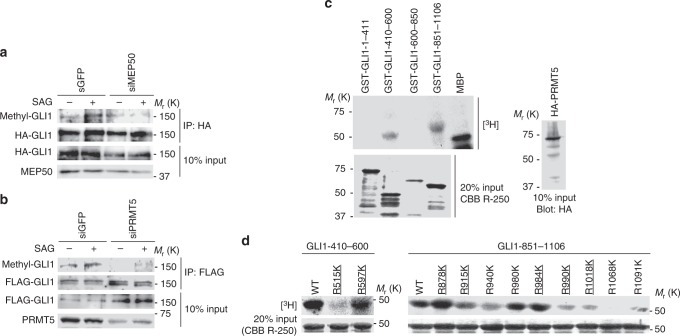
Fig. 4.
MEP50/PRMT5 complex induces GLI1 methylation. a, b Methylation of GLI1 in MEP50- (a) or PRMT5- (b) knockdown C3H10T1/2 cells. siMEP50-m2 and siPRMT5-m2 siRNAs were stably expressed by recombinant retroviruses. Cells transfected with FLAG-GLI1 were cultured for 24 h, followed by treatment with 300 nM SAG for 24 h. Methylated GLI1 was detected by immunoprecipitation with an anti-FLAG antibody followed by immunoblot with anti-SYM11 antibody. c In vitro methylation assays to determine the region including methylated arginine residues in GLI1 deletion mutants. HA-PRMT5 expression plasmid was transfected into HEK293T cells. At 48 h after transfection, the cells were lysed, and HA-PRMT5 was immunoprecipitated using an anti-HA (3F10) antibody. GST-GLI1 deletion mutants coupled to glutathione sepharose were incubated with immunoprecipitated HA-PRMT5 from HEK293T cells. Upper panel represents the methylated GST-GLI1 deletion mutant. Lower panel represents 20% input of GST-GLI1 deletion mutants detected by CBB R-250 staining. HA-PRMT5 expressed in 10% of total lysate used for immunoprecipitation is shown in the right panel. d In vitro methylation assays to determine methylation sites in GLI1 using amino acid substitutions (arginine to lysine) of candidate methylation sites. In vitro methylation assays were performed as described in (c). Upper panel represents methylated GST-GLI1 mutants. Lower panel represents 20% input of GST-GLI1 mutants detected by CBB R-250 staining. Underlined text denotes highly conserved residues among mammals, as shown in Supplementary Fig. 4. In c, data represent one of three independent experiments with similar results. In a and d, data represent one of twice independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Fig. 6